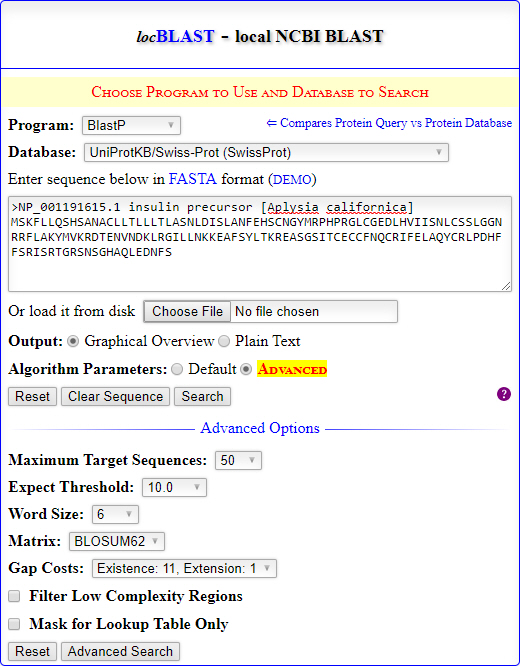
locBLAST is a PHP library for enriching command-line NCBI BLAST+ programs to an interactive graphical user interface. It performs both local and remote database search through a PHP supported web server. The PHP library executes the command-line NCBI BLAST+ programs using exec() function by passing parameters from the HTML form fields. In PHP, the system(), exec(), passthru(), and shell_exec() functions are used to pass system commands.
The locBLAST allows users to input query sequence by pasting in the text box or file uploading. Currently, locBLAST supports nine BLAST+ programs such as blastn, blastp, blastx, tblastn, tblastx, deltablast, psiblast, rpsblast, and rpstblastn for sequence search. Moreover, the latest release locBLAST supports various platforms such as Windows, Linux, and MacOS. The locBLAST PHP library and test database files were freely available at GitHub.
The implementation of locBLAST in a web server is very easy. It requires standalone NCBI BLAST+ suite and locBLAST library. The latest release of NCBI BLAST+ (as on Nov 17, 2020, the stable release is 2.11.0) can be downloaded from the NCBI FTP server (ftp://ftp.ncbi.nih.gov/blast/executables/blast+/LATEST). A simple PHP supporting web server is enough to run the BLAST+ program through the web browser.
A simple protocol to setup the locBLAST is given below,
- Download the compressed NCBI BLAST+ suite from the NCBI FTP server (ftp://ftp.ncbi.nih.gov/blast/executables/blast+/LATEST/ncbi-blast-2.7.1+-x64-linux.tar.gz, for Linux 64-bit operating system).
- Create a directory named locBLAST (optional) in the parent web directory (usually named htdocs, www, wwwroot, or webpath).
- Decompress the ncbi-blast-2.11.0+-x64-linux.tar.gz file and copy all the files present in the bin folder to locBLAST directory.
- Download locBLAST library from the GitHub and copy to locBLAST directory.
- Open the URL http://localhost/locBLAST in the web browser to run BLAST+ programs.
- Click the hyperlink DEMO and Search button to test local database search. The default parameters will generate an graphical search report containing graphical color key, tabular summary, and formatted alignment.
The form fields in locBLAST library can be extended or modified with custom form fields to perform advanced search. Moreover, desired database files of biological databases such as PDB, UniProtKB/Swiss-Prot, EST, RefSeq., etc. can be downloaded from the NCBI FTP directory (ftp://ftp.ncbi.nih.gov/blast/db/) and copied to the db directory to perform offline database search.
We can create custom database using makeblastdb (for nucleotide or protein) or makeprofiledb (for profiles) program present in the locBLAST directory. To create a custom database, copy the FASTA file formatted multiple sequence file to the db directory. Then run the command ./makeblastdb -in db/pdt.fas -out db/pdt -dbtype prot -title PDTDB directing to the locBLAST directory to create a protein custom database named PDTDB (already included in the db folder).
A model FASTA file formatted multiple sequence file pdt.fas is below:
>pdt|pdtdbt00001|pdb|1OKE|A/B
MRCIGISNRDFVEGVSGGSWVDIVLEHGSCVTTMAKNKPTLDFELIKTEAKQPATLRKYC
IEAKLTNTTTESRCPTQGEPTLNEEQDKRFVCKHSMVDRGWGNGCGLFGKGGIVTCAMFT
CKKNMEGKIVQPENLEYTVVITPHSGEEHAVGNDTGKHGKEVKITPQSSITEAELTGYGT
VTMECSPRTGLDFNEMVLLQMKDKAWLVHRQWFLDLPLPWLPGADTQGSNWIQKETLVTF
KNPHAKKQDVVVLGSQEGAMHTALTGATEIQMSSGNLLFTGHLKCRLRMDKLQLKGMSYS
MCTGKFKVVKEIAETQHGTIVIRVQYEGDGSPCKIPFEIMDLEKRHVLGRLITVNPIVTE
KDSPVNIEAEPPFGDSYIIIGVEPGQLKLNWFKK
>pdt|pdtdbt00002|pdb|4O6B|A/B
AHHHHHHSSGVDLGTENLYFQSNADSGCVVSWKNKELKCGSGIFITDNVHTWTEQYKFQP
ESPSKLASAIQKAHEEGICGIRSVTRLENLMWKQITPELNHILSENEVKLTIMTGDIKGI
MQAGKRSLRPQPTELKYSWKTWGKAKMLSTESHNQTFLIDGPETAECPNTNRAWNSLEVE
DYGFGVFTTNIWLKLKEKQDVFCDSKLMSAAIKDNRAVHADMGYWIESALNDTWKIEKAS
FIEVKNCHWPKSHTLWSNGVLESEMIIPKNLAGPVSQHNYRPGYHTQITGPWHLGKLEMD
FDFCDGTTVVVTEDCGNRGPSLRTTTASGKLITEWCCRSCTLPPLRYRGEDGCWYGMEIR
PLKEKEENLVNSLVTA
>pdt|pdtdbt00003|pdb|2IOK|A/B
SKKNSLALSLTADQMVSALLDAEPPILYSEYDPTRPFSEASMMGLLTNLADRELVHMINW
AKRVPGFVDLTLHDQVHLLECAWLEILMIGLVWRSMEHPGKLLFAPNLLLDRNQGKCVEG
MVEIFDMLLATSSRFRMMNLQGEEFVCLKSIILLNSGVYTFLSSTLKSLEEKDHIHRVLD
KITDTLIHLMAKAGLTLQQQHQRLAQLLLILSHIRHMSNKGMEHLYSMKCKNVVPLYDLL
LEMLDAHRLHAPTS
>pdt|pdtdbt00004|pdb|2Q1Y|A/B
MTPPHNYLAVIKVVGIGGGGVNAVNRMIEQGLKGVEFIAINTDAQALLMSDADVKLDVGR
DSTRGLGAGADPEVGRKAAEDAKDEIEELLRGADMVFVTAGEGGGTGTGGAPVVASIARK
LGALTVGVVTRPFSFEGKRRSNQAENGIAALRESCDTLIVIPNDRLLQMGDAAVSLMDAF
RSADEVLLNGVQGITDLITTPGLINVDFADVKGIMSGAGTALMGIGSARGEGRSLKAAEI
AINSPLLEASMEGAQGVLMSIAGGSDLGLFEINEAASLVQDAAHPDANIIFGTVIDDSLG
DEVRVTVIAAGFDVSGPGRKPVMGETGGAHRIESAKAGKLTSTLFEPVDAVSVPLHTNGA
TLSIGGDDDDVDVPPFMRR
>pdt|pdtdbt00005|pdb|3GGE|A/B/C
SMKGIEKEVNVYKSEDSLGLTITDNGVGYAFIKRIKDGGVIDSVKTICVGDHIESINGEN
IVGWRHYDVAKKLKELKKEELFTMKLIEPKKSSEA
>pdt|pdtdbt00006|pdb|2F9Q|A/B/C/D
MAKKTSSKGKLPPGPLPLPGLGNLLHVDFQNTPYCFDQLRRRFGDVFSLQLAWTPVVVLN
GLAAVREALVTHGEDTADRPPVPITQILGFGPRSQGVFLARYGPAWREQRRFSVSTLRNL
GLGKKSLEQWVTEEAACLCAAFANHSGRPFRPNGLLDKAVSNVIASLTCGRRFEYDDPRF
LRLLDLAQEGLKEESGFLREVLNAVPVDRHIPALAGKVLRFQKAFLTQLDELLTEHRMTW
DPAQPPRDLTEAFLAEMEKAKGNPESSFNDENLRIVVADLFSAGMVTTSTTLAWGLLLMI
LHPDVQRRVQQEIDDVIGQVRRPEMGDQAHMPYTTAVIHEVQRFGDIVPLGMTHMTSRDI
EVQGFRIPKGTTLITNLSSVLKDEAVWEKPFRFHPEHFLDAQGHFVKPEAFLPFSAGRRA
CLGEPLARMELFLFFTSLLQHFSFSVPTGQPRPSHHGVFAFLVSPSPYELCAVPRHHHH
>pdt|pdtdbt00007|pdb|3EAH|A/B
PKFPRVKNWEVGSITYDTLSAQAQQDGPCTPRRCLGSLVFPRKLQGRPSPGPPAPEQLLS
QARDFINQYYSSIKRSGSQAHEQRLQEVEAEVAATGTYQLRESELVFGAKQAWRNAPRCV
GRIQWGKLQVFDARDCRSAQEMFTYICNHIKYATNRGNLRSAITVFPQRCPGRGDFRIWN
SQLVRYAGYRQQDGSVRGDPANVEITELCIQHGWTPGNGRFDVLPLLLQAPDEPPELFLL
PPELVLEVPLEHPTLEWFAALGLRWYALPAVSNMLLEIGGLEFPAAPFSGWYMSTEIGTR
NLCDPHRYNILEDVAVCMDLDTRTTSSLWKDKAAVEINVAVLHSYQLAKVTIVDHHAATA
SFMKHLENEQKARGGCPADWAWIVPPISGSLTPVFHQEMVNYFLSPAFRYQPDPWKGSAA
KGTGITR
>pdt|pdtdbt00008|pdb|4PQE|A
EGREDAELLVTVRGGRLRGIRLKTPGGPVSAFLGIPFAEPPMGPRRFLPPEPKQPWSGVV
DATTFQSVCYQYVDTLYPGFEGTEMWNPNRELSEDCLYLNVWTPYPRPTSPTPVLVWIYG
GGFYSGASSLDVYDGRFLVQAERTVLVSMNYRVGAFGFLALPGSREAPGNVGLLDQRLAL
QWVQENVAAFGGDPTSVTLFGESAGAASVGMHLLSPPSRGLFHRAVLQSGAPNGPWATVG
MGEARRRATQLAHLVGCPPGGTGGNDTELVACLRTRPAQVLVNHEWHVLPQESVFRFSFV
PVVDGDFLSDTPEALINAGDFHGLQVLVGVVKDEGSYFLVYGAPGFSKDNESLISRAEFL
AGVRVGVPQVSDLAAEAVVLHYTDWLHPEDPARLREALSDVVGDHNVVCPVAQLAGRLAA
QGARVYAYVFEHRASTLSWPLWMGVPHGYEIEFIFGIPLDPSRNYTAEEKIFAQRLMRYW
ANFARTGDPNEPRDPKAPQWPPYTAGAQQYVSLDLRPLEVRRGLRAQACAFWNRFLPKLL
SAT
The custom profile/CDD database can be created using command ./makeprofiledb -in db/CDD/Smart.pn -out db/CDD/Smart -dbtype rps -title SMART.v6.0 for psiblast, rpsblast, and rpstblastn programs, and command ./makeprofiledb -in db/CDD/cdd_delta.pn -out db/CDD/cdd_delta -dbtype delta -title cdd_delta for deltablast program.
The current release locBLAST v2.1 has added list of features below,
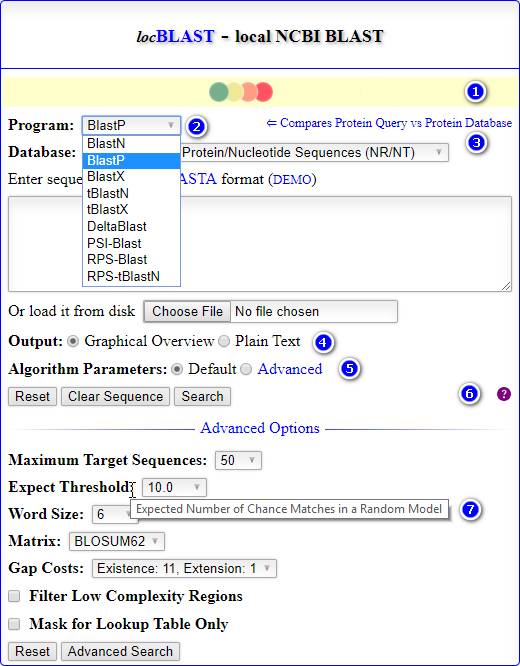
- The home page of locBLAST v2.1 web form has added with more features, (1) progress bar and status bar, (2) advanced BLAST+ programs
deltablast,psiblast,rpsblast, andrpstblastn, (3) hyperlink to detailed program description page, with short title, (4) graphical or plain text output, (5) advanced algorithm parameters, (6) tutorial page & BLAST release check, and (7) mouse over tooltip text description of the keyword.
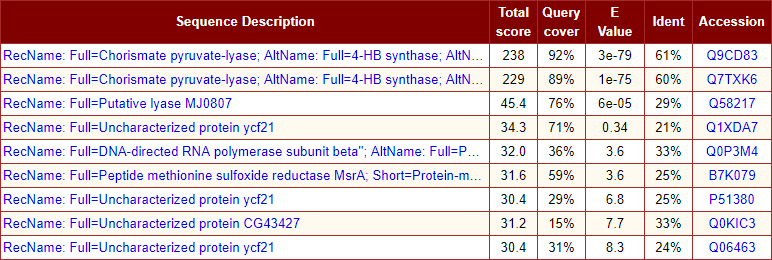
- Graphical overview of the sequence alignment hits with mouse over tooltip text description on hyperlinks. Color keys represents the score of the sequence alignment.
- Summary table of sequence alignment hits consisting of sequence description, total score, percentage of query coverage, e-value, identity, and accession number. Hyperlinks to alignment hit section and reference link to biological sequence database.
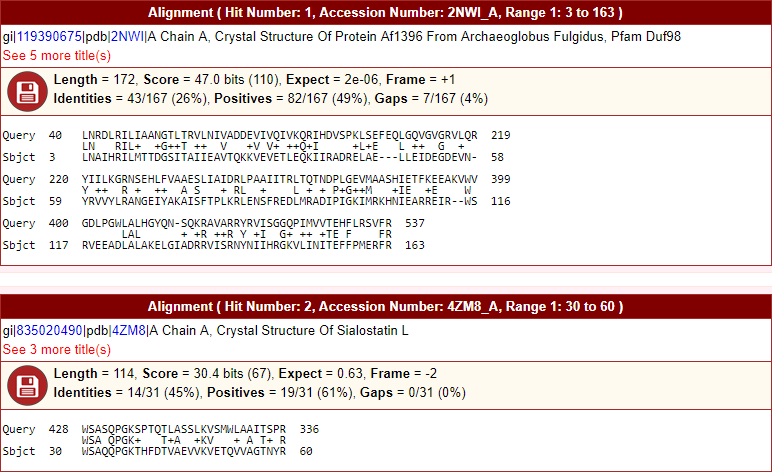
- Formatted sequence alignment for plus, minus, six-reading frames, and translated nucleotide sequences. Hyperlinks to biological sequence database in header description. Hide or show (toggle) options to large descriptions. Allows to save sequence to the disk in FASTA file format.
- Latest NCBI BLAST+ binary/executables release check.
- Simple tutorial on locBLAST usage.
The snapshot of graphical output is here.
The snapshot of plain text output is here.
Please feel free to sent your queries, suggestions and/or comments related to locBLAST program to [email protected] or [email protected].
locBLAST is made available under version 3 of the GNU Lesser General Public License.
Ashok Kumar, T. (2019). locBLAST v2.0 - an improved PHP library for embedding standalone NCBI BLAST+ program to an interactive graphical user interface. bioRxiv. [Abstract] [PDF]