diff --git a/README.md b/README.md
index 95b36e9a..2cbb2e92 100644
--- a/README.md
+++ b/README.md
@@ -36,7 +36,9 @@ The following figure is the simplified overview of Geochemistry π:
The following figure is the frontend-backend separation architecture of Geochemistry:
-
+
+

+
The following figure is the customized automated ML pipeline:
-
+
+

+
@@ -158,7 +162,9 @@ The following figure is the design pattern hierarchical architecture:
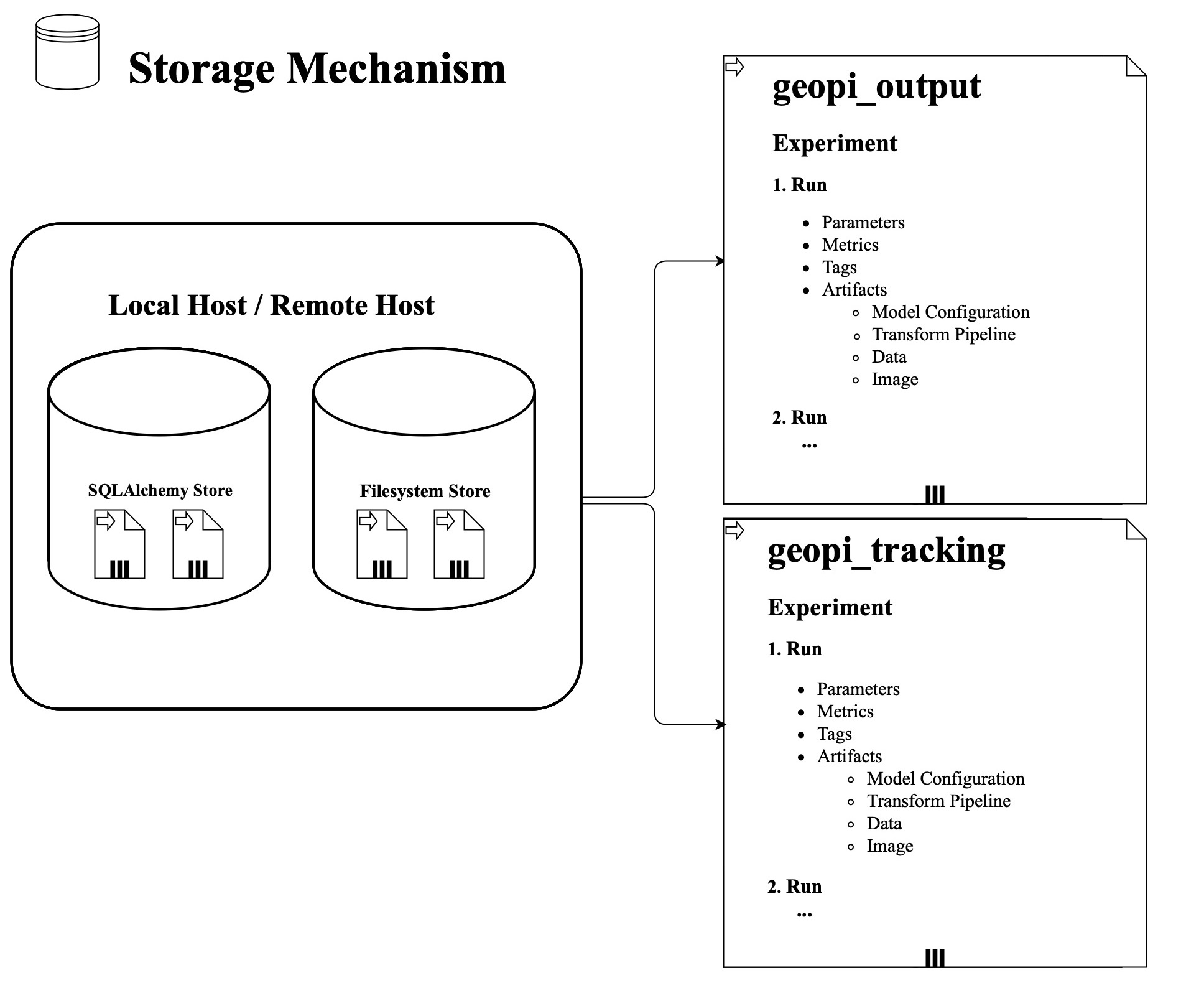
The following figure is the storage mechanism:
-
+
+

+