From 37d91addf48aee59b366a87f08a8302d3bc6cb55 Mon Sep 17 00:00:00 2001
From: jin <571979568@qq.com>
Date: Mon, 25 Dec 2023 17:23:59 +0800
Subject: [PATCH 1/4] docs:Added A new document-Add New Model To Framework.md &
Added new content to decomposition.md document
---
.../Add New Model To Framework.md | 377 ++++++++++++
.../Decomposition/decomposition.md | 544 ++++++++++++++++++
docs/source/conf.py | 13 +-
docs/source/index.rst | 2 +-
.../geochemistrypi.data_mining.data.rst | 8 +
5 files changed, 937 insertions(+), 7 deletions(-)
create mode 100644 docs/source/For Developer/Add New Model To Framework.md
diff --git a/docs/source/For Developer/Add New Model To Framework.md b/docs/source/For Developer/Add New Model To Framework.md
new file mode 100644
index 00000000..9e60ac82
--- /dev/null
+++ b/docs/source/For Developer/Add New Model To Framework.md
@@ -0,0 +1,377 @@
+
+# Add New Model To Framework
+
+
+
+
+## Table of Contents
+
+- [1. Understand the model](#1-understand-the-model)
+- [2. Add Model](#2-add-model)
+ - [2.1 Add The Model Class](#21-add-the-model-class)
+ - [2.1.1 Find Add File](#211-find-add-file)
+ - [2.1.2 Define class properties and constructors, etc.](#212-define-class-properties-and-constructors-etc)
+ - [2.1.3 Define manual\_hyper\_parameters](#213-define-manual_hyper_parameters)
+ - [2.1.4 Define special\_components](#214-define-special_components)
+ - [2.2 Add AutoML](#22-add-automl)
+ - [2.2.1 Add AutoML code to class](#221-add-automl-code-to-class)
+ - [2.3 Get the hyperparameter value through interactive methods](#23-get-the-hyperparameter-value-through-interactive-methods)
+ - [2.3.1 Find file](#231-find-file)
+ - [2.3.2 Create the .py file and add content](#232-create-the-py-file-and-add-content)
+ - [2.3.3 Import in the file that defines the model class](#233-import-in-the-file-that-defines-the-model-class)
+ - [2.4 Call Model](#24-call-model)
+ - [2.4.1 Find file](#241-find-file)
+ - [2.4.2 Import module](#242-import-module)
+ - [2.4.3 Call model](#243-call-model)
+ - [2.5 Add the algorithm list and set NON\_AUTOML\_MODELS](#25-add-the-algorithm-list-and-set-non_automl_models)
+ - [2.5.1 Find file](#251-find-file)
+- [3. Test model](#3-test-model)
+- [4. Completed Pull Request](#4-completed-pull-request)
+- [5. Precautions](#5-precautions)
+
+
+## 1. Understand the model
+You need to understand the general meaning of the model, determine which algorithm the model belongs to and the role of each parameter.
++ You can choose to learn about the relevant knowledge on the [scikit-learn official website](https://scikit-learn.org/stable/index.html).
+
+
+## 2. Add Model
+### 2.1 Add The Model Class
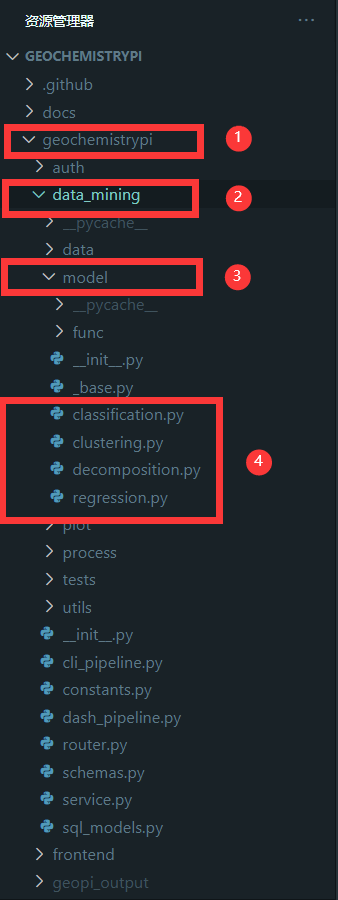
+#### 2.1.1 Find Add File
+First, you need to define the model class that you need to complete in the corresponding algorithm file. The corresponding algorithm file is in the `model` folder in the `data_mining` folder in the `geochemistrypi` folder.
+
+
+
+**eg:** If you want to add a model to the regression algorithm, you need to add it in the `regression.py` file.
+
+
+#### 2.1.2 Define class properties and constructors, etc.
+(1) Define the class and the required Base class
+```
+class NAME (Base class):
+```
++ NAME is the name of the algorithm, you can refer to the NAME of other models, the format needs to be consistent.
++ Base class needs to be selected according to the actual algorithm requirements.
+
+```
+"""The automation workflow of using "Name" to make insightful products."""
+```
++ Class explanation, you can refer to other classes.
+
+(2) Define the name and the special_function
+
+```
+name = "name"
+```
++ Define name, different from NMAE.
++ This name needs to be added to the _`constants.py`_ file and the corresponding algorithm file in the `process` folder. Note that the names are consistent.
+```
+special_function = []
+```
++ special_function is added according to the specific situation of the model, you can refer to other similar models.
+
+(3) Define constructor
+```
+def __init__(
+ self,
+ parameter:type=Default parameter value,
+ ) -> None:
+```
++ All parameters in the corresponding model function need to be written out.
++ Default parameter value needs to be set according to official documents.
+
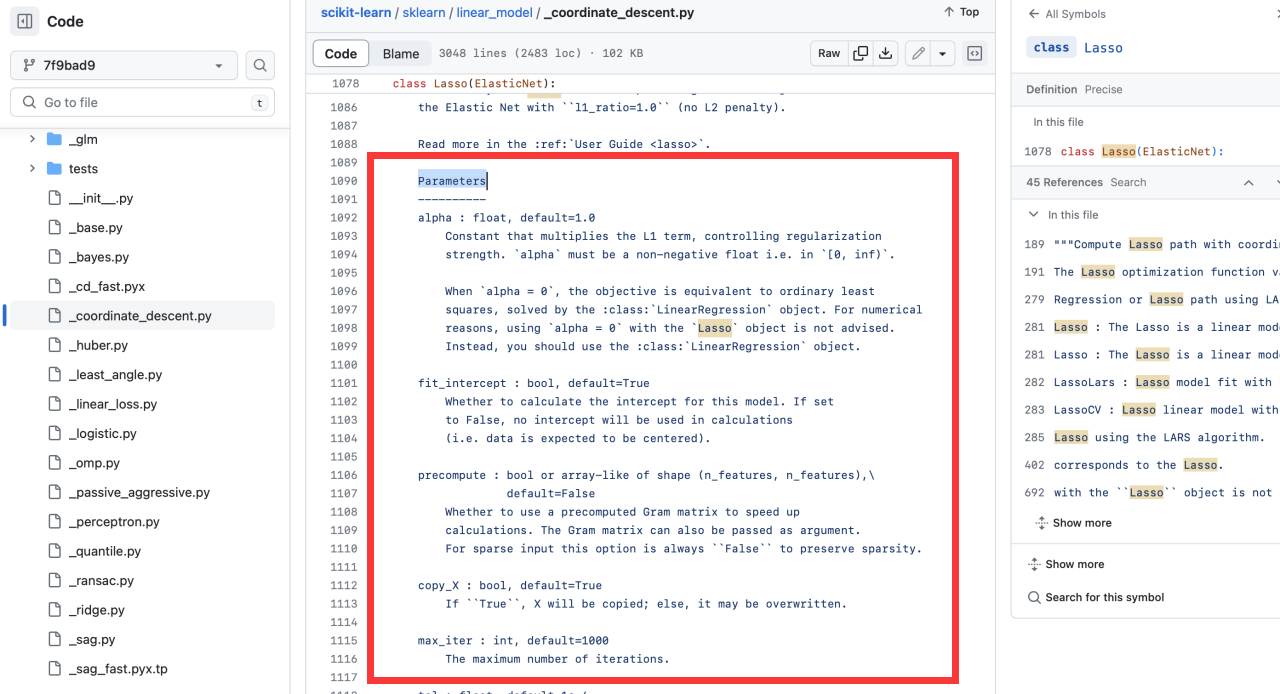
+```
+ """
+Parameters
+----------
+parameter:type,default = Dedault
+
+References
+----------
+Scikit-learn API: sklearn.model.name
+https://scikit-learn.org/......
+```
++ Parameters is in the source of the corresponding model on the official website of sklearn
+
+**eg:** Take the [Lasso algorithm](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Lasso.html) as a column.
+
+
+
+
+
++ References is your model's official website.
+
+(4) The constructor of Base class is called
+```
+super().__init__()
+```
+(5) Initializes the instance's state by assigning the parameter values passed to the constructor to the instance's properties.
+```
+self.parameter=parameter
+```
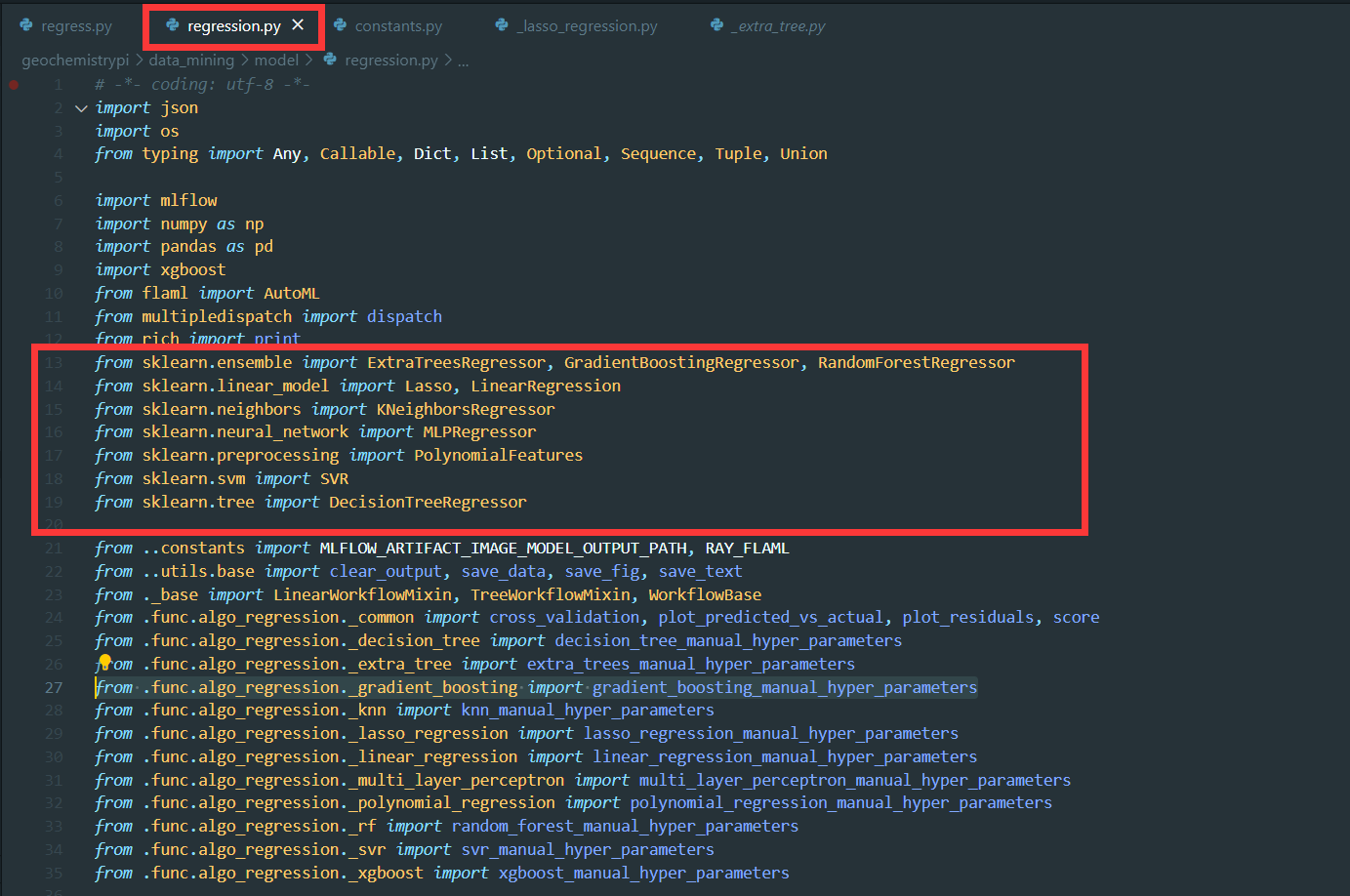
+(6) Create the model and assign
+```
+self.model=modelname(
+ parameter=self.parameter
+)
+```
+**Note:** Don't forget to import Model from scikit-learn
+
+
+
+(7) Define other class properties
+```
+self.properties=...
+```
+
+#### 2.1.3 Define manual_hyper_parameters
+manual_hyper_parameters gets the hyperparameter value by calling the manual hyperparameter function, and returns hyper_parameters.
+```
+hyper_parameters = name_manual_hyper_parameters()
+```
++ This function calls the corresponding function in the `func` folder (needs to be written, see 2.2.2) to get the hyperparameter value.
+
++ This function is called in the corresponding file of the `Process` folder (need to be written, see 2.3).
++ Can be written with reference to similar classes
+
+
+#### 2.1.4 Define special_components
+Its purpose is to Invoke all special application functions for this algorithms by Scikit-learn framework.
+**Note:** The content of this part needs to be selected according to the actual situation of your own model.Can refer to similar classes.
+
+```
+GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH")
+```
++ This line of code gets the image model output path from the environment variable.
+```
+GEOPI_OUTPUT_ARTIFACTS_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_PATH")
+```
++ This line of code takes the general output artifact path from the environment variable.
+**Note:** You need to choose to add the corresponding path according to the usage in the following functions.
+
+### 2.2 Add AutoML
+#### 2.2.1 Add AutoML code to class
+(1) Set AutoML related parameters
+```
+ @property
+ def settings(self) -> Dict:
+ """The configuration of your model to implement AutoML by FLAML framework."""
+ configuration = {
+ "time_budget": '...'
+ "metric": '...',
+ "estimator_list": '...'
+ "task": '...'
+ }
+ return configuration
+```
++ "time_budget" represents total running time in seconds
++ "metric" represents Running metric
++ "estimator_list" represents list of ML learners
++ "task" represents task type
+**Note:** You can keep the parameters consistent, or you can modify them to make the AutoML better.
+
+(2) Add parameters that need to be AutoML
+You can add the parameter tuning code according to the following code:
+```
+ @property
+ def customization(self) -> object:
+ """The customized 'Your model' of FLAML framework."""
+ from flaml import tune
+ from flaml.data import 'TPYE'
+ from flaml.model import SKLearnEstimator
+ from sklearn.ensemble import 'model_name'
+
+ class 'Model_Name'(SKLearnEstimator):
+ def __init__(self, task=type, n_jobs=None, **config):
+ super().__init__(task, **config)
+ if task in 'TOYE':
+ self.estimator_class = 'model_name'
+
+ @classmethod
+ def search_space(cls, data_size, task):
+ space = {
+ "'parameters1'": {"domain": tune.uniform(lower='...', upper='...'), "init_value": '...'},
+ "'parameters2'": {"domain": tune.choice([True, False])},
+ "'parameters3'": {"domain": tune.randint(lower='...', upper='...'), "init_value": '...'},
+ }
+ return space
+
+ return "Model_Name"
+```
+**Note1:** The content in ' ' needs to be modified according to your specific code
+**Note2:**
+```
+ space = {
+ "'parameters1'": {"domain": tune.uniform(lower='...', upper='...'), "init_value": '...'},
+ "'parameters2'": {"domain": tune.choice([True, False])},
+ "'parameters3'": {"domain": tune.randint(lower='...', upper='...'), "init_value": '...'},
+ }
+```
+
++ tune.Uniform represents float
++ tune.choice represents bool
++ tune.randint represents int
++ lower represents the minimum value of the range, upper represents the maximum value of the range, and init_value represents the initial value
+**Note:** You need to select parameters based on the actual situation of the model
+
+(3) Define special_components(FLAML)
+This part is the same as 2.1.4 as a whole, and can be modified with reference to it, but only the following two points need to be noted:
+a.The multi-dispatch function is different
+Scikit-learn framework:@dispatch()
+FLAML framework:@dispatch(bool)
+
+b.Added 'is_automl: bool' to the def
+**eg:**
+```
+Scikit-learn framework:
+def special_components(self, **kwargs) -> None:
+
+FLAML framework:
+def special_components(self, is_automl: bool, **kwargs) -> None:
+```
+c.self.model has a different name
+**eg:**
+```
+Scikit-learn framework:
+coefficient=self.model.coefficient
+
+FLAML framework:
+coefficient=self.auto_model.coefficient
+```
+
+**Note:** You can refer to other similar codes to complete your code.
+
+### 2.3 Get the hyperparameter value through interactive methods
+Sometimes the user wants to modify the hyperparameter values for model training, so you need to establish an interaction to get the user's modifications.
+
+
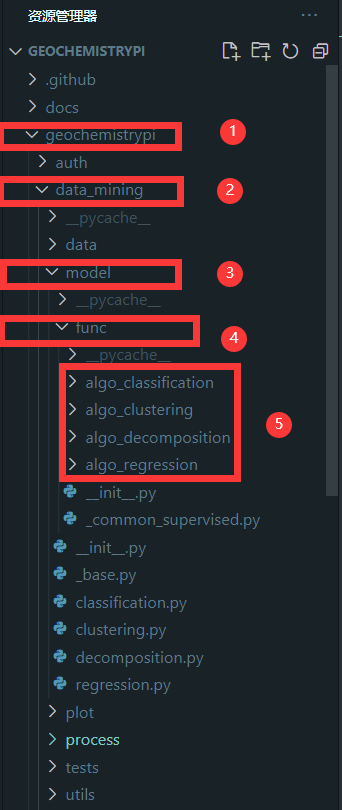
+#### 2.3.1 Find file
+You need to find the corresponding folder for model. The corresponding algorithm file is in the `func` folder in the model folder in the `data_mining` folder in the `geochemistrypi` folder.
+
+
+
+**eg:** If your model belongs to the regression, you need to add the `corresponding.py` file in the `alog_regression` folder.
+
+
+#### 2.3.2 Create the .py file and add content
+(1) Create a .py file
+**Note:** Keep name format consistent.
+
+(2) Import module
+```
+from typing import Dict
+from rich import print
+from ....constants import SECTION
+```
++ In general, these modules need to be imported
+```
+from ....data.data_readiness import bool_input, float_input, num_input
+```
++ This needs to choose the appropriate import according to the hyperparameter type of model interaction.
+
+(3) Define the function
+```
+def name_manual_hyper_parameters() -> Dict:
+```
+**Note:** The name needs to be consistent with that in 2.1.3.
+
+(4) Interactive format
+```
+print("Hyperparameters: Role")
+print("Recommended value")
+Hyperparameters = type_input(Recommended value, SECTION[2], "@Hyperparameters: ")
+```
+**Note:** The recommended value needs to be the default value of the corresponding package.
+
+(5) Integrate all hyperparameters into a dictionary type and return.
+```
+hyper_parameters = {
+ "Hyperparameters1": Hyperparameters1,
+ "Hyperparameters": Hyperparameters2,
+}
+retuen hyper_parameters
+```
+
+#### 2.3.3 Import in the file that defines the model class
+```
+from .func.algo_regression.Name import name_manual_hyper_parameters
+```
+**eg:**
+
+
+### 2.4 Call Model
+
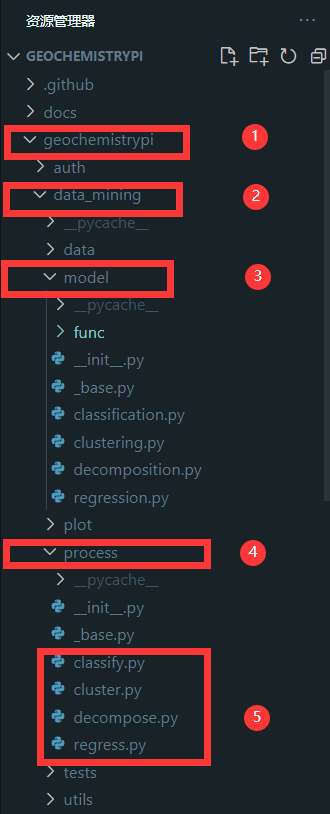
+#### 2.4.1 Find file
+Call the model in the corresponding file in the `process` folder. The corresponding algorithm file is in the `process` folder in the` model` folder in the `data_mining` folder in the `geochemistrypi` folder.
+
+
+
+**eg:** If your model belongs to the regression,you need to call it in the regress.py file.
+
+#### 2.4.2 Import module
+You need to add your model in the from ..model.regression import().
+```
+from ..model.regression import(
+ ...
+ NAME,
+)
+```
+**Note:** NAME needs to be the same as the NAME when defining the class in step 2.1.2.
+**eg:**
+
+
+
+#### 2.4.3 Call model
+There are two activate methods defined in the Regression and Classification algorithms, the first method uses the Scikit-learn framework, and the second method uses the FLAML and RAY frameworks. Decomposition and Clustering algorithms only use the Scikit-learn framework. Therefore, in the call, Regression and Classification need to add related codes to implement the call in both methods, and only one time is needed in Clustering and Decomposition.
+
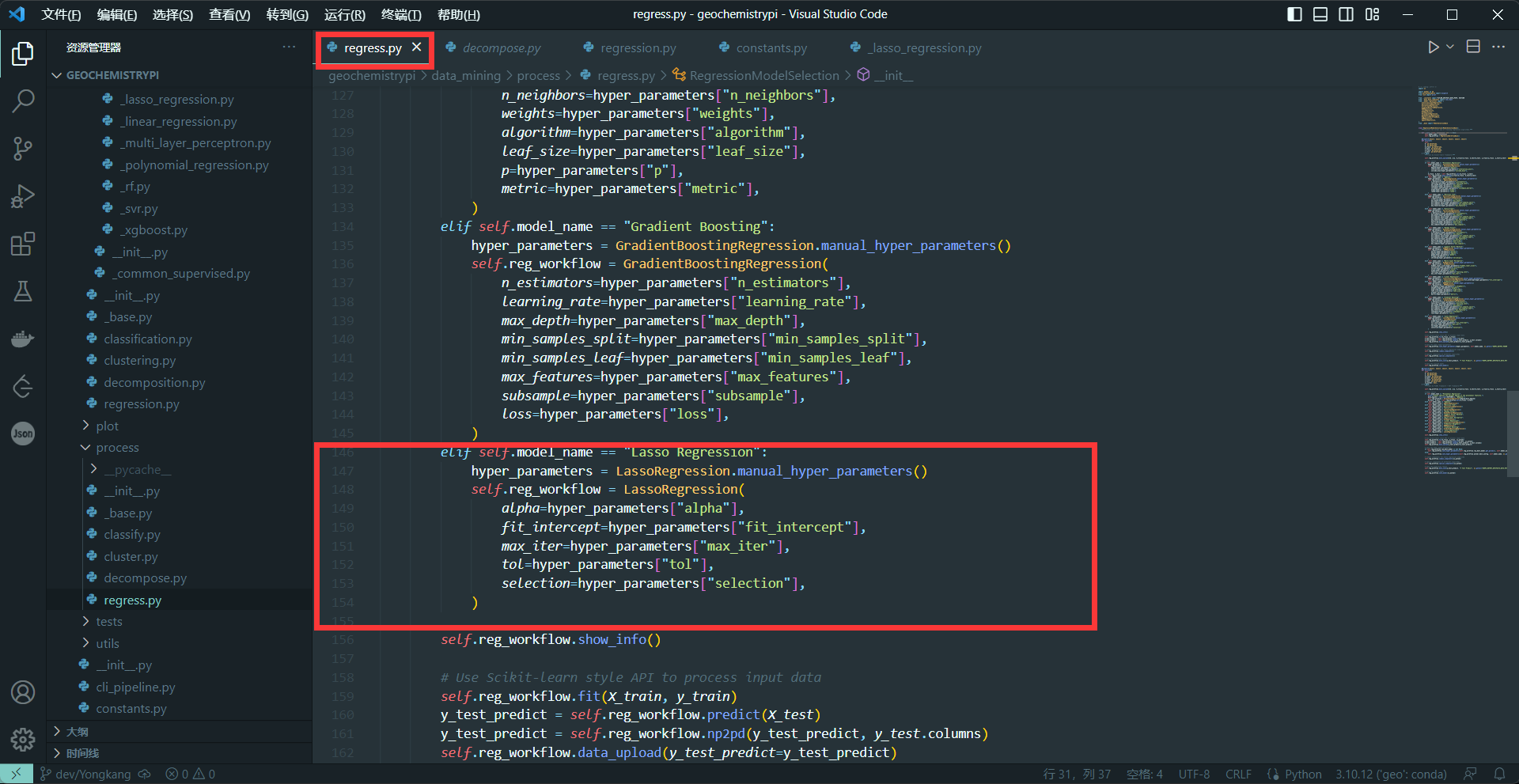
+(1) Call model in the first activate method(Including Classification, Regression,Decomposition,Clustering)
+```
+elif self.model_name == "name":
+ hyper_parameters = NAME.manual_hyper_parameters()
+ self.dcp_workflow = NAME(
+ Hyperparameters1=hyper_parameters["Hyperparameters2"],
+ Hyperparameters1=hyper_parameters["Hyperparameters2"],
+ ...
+ )
+```
++ The name needs to be the same as the name in 2.4
++ The hyperparameters in NAME() are the hyperparameters obtained interactively in 2.2
+**eg:**
+
+
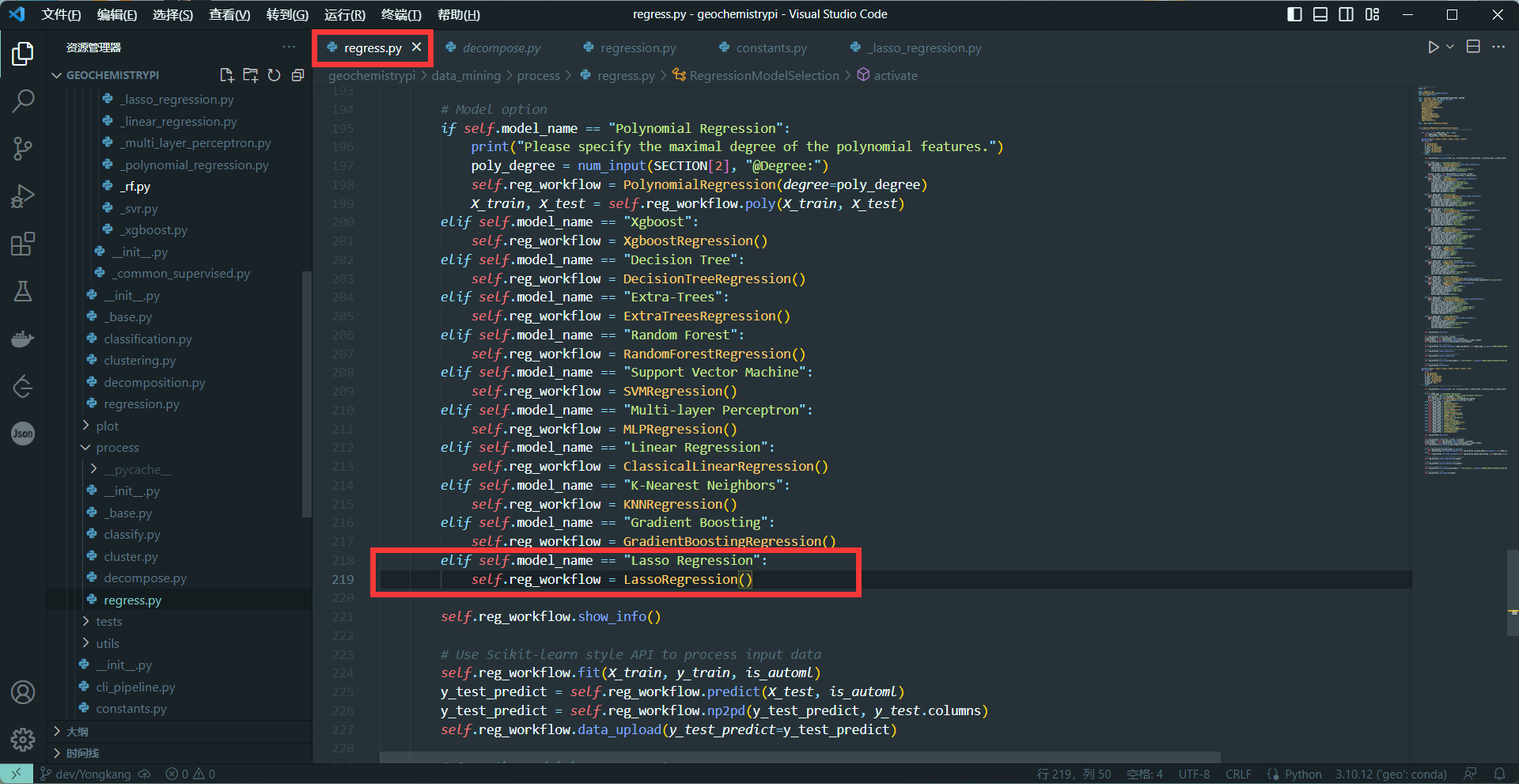
+(2)Call model in the second activate method(Including Classification, Regression)
+```
+elif self.model_name == "name":
+ self.reg_workflow = NAME()
+```
++ The name needs to be the same as the name in 2.4
+**eg:**
+
+
+### 2.5 Add the algorithm list and set NON_AUTOML_MODELS
+
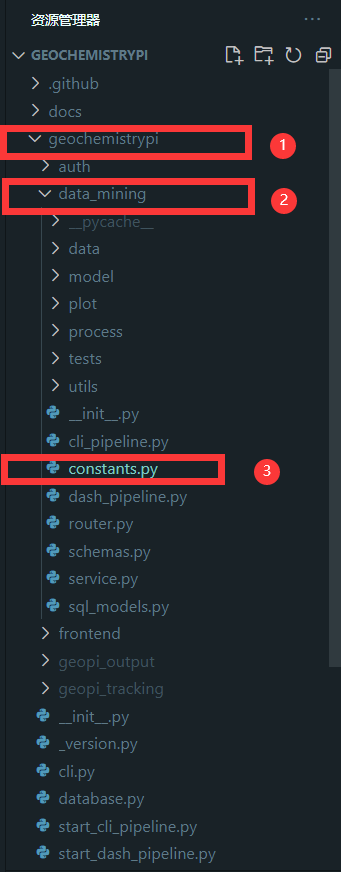
+#### 2.5.1 Find file
+Find the constants file to add the model name,The constants file is in the `data_mining` folder in the `geochemistrypi` folder.
+
+
+
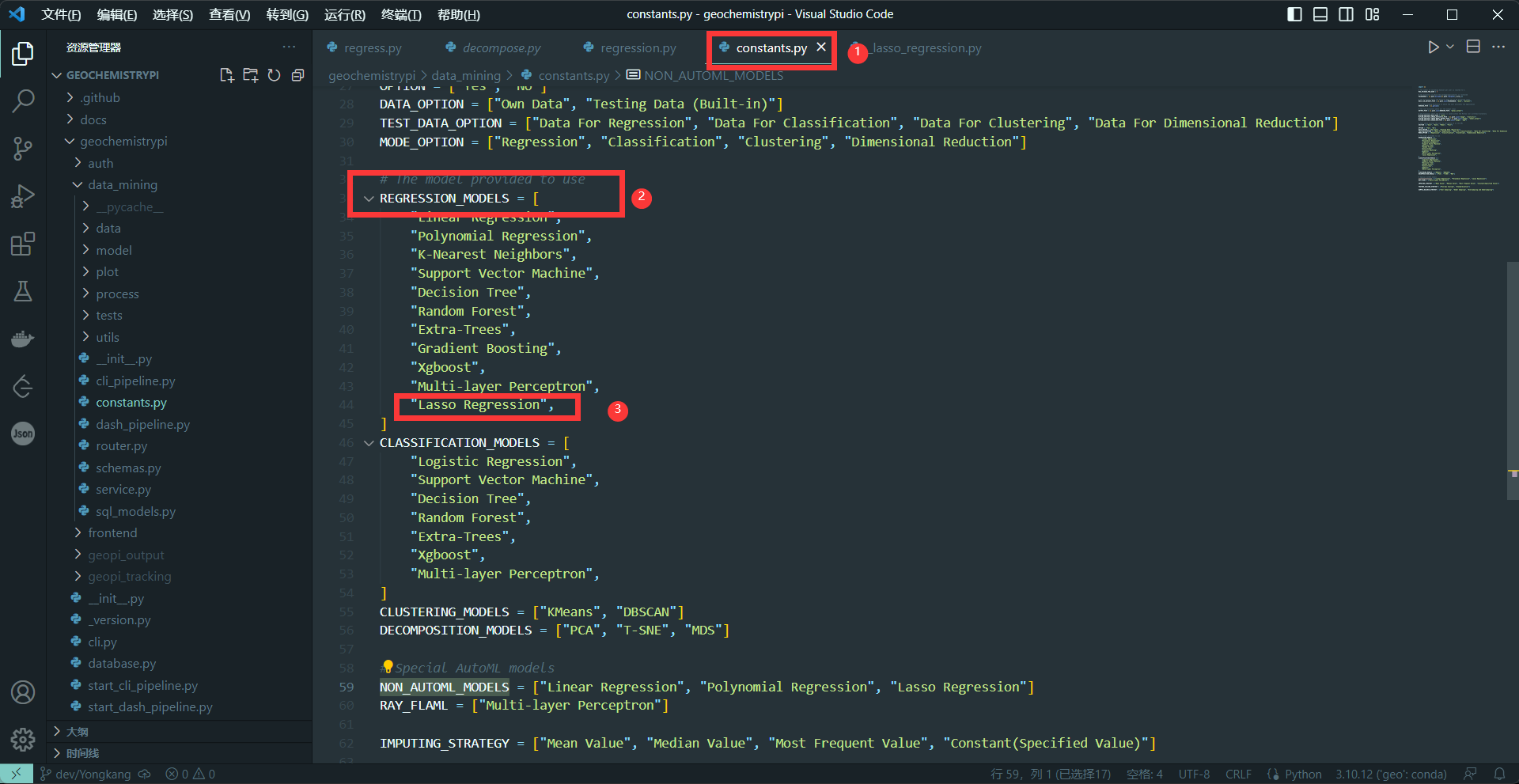
+(1) Add the model name
+Add model name to the algorithm list corresponding to the model in the constants file.
+**eg:** Add the name of the Lasso regression algorithm.
+
+
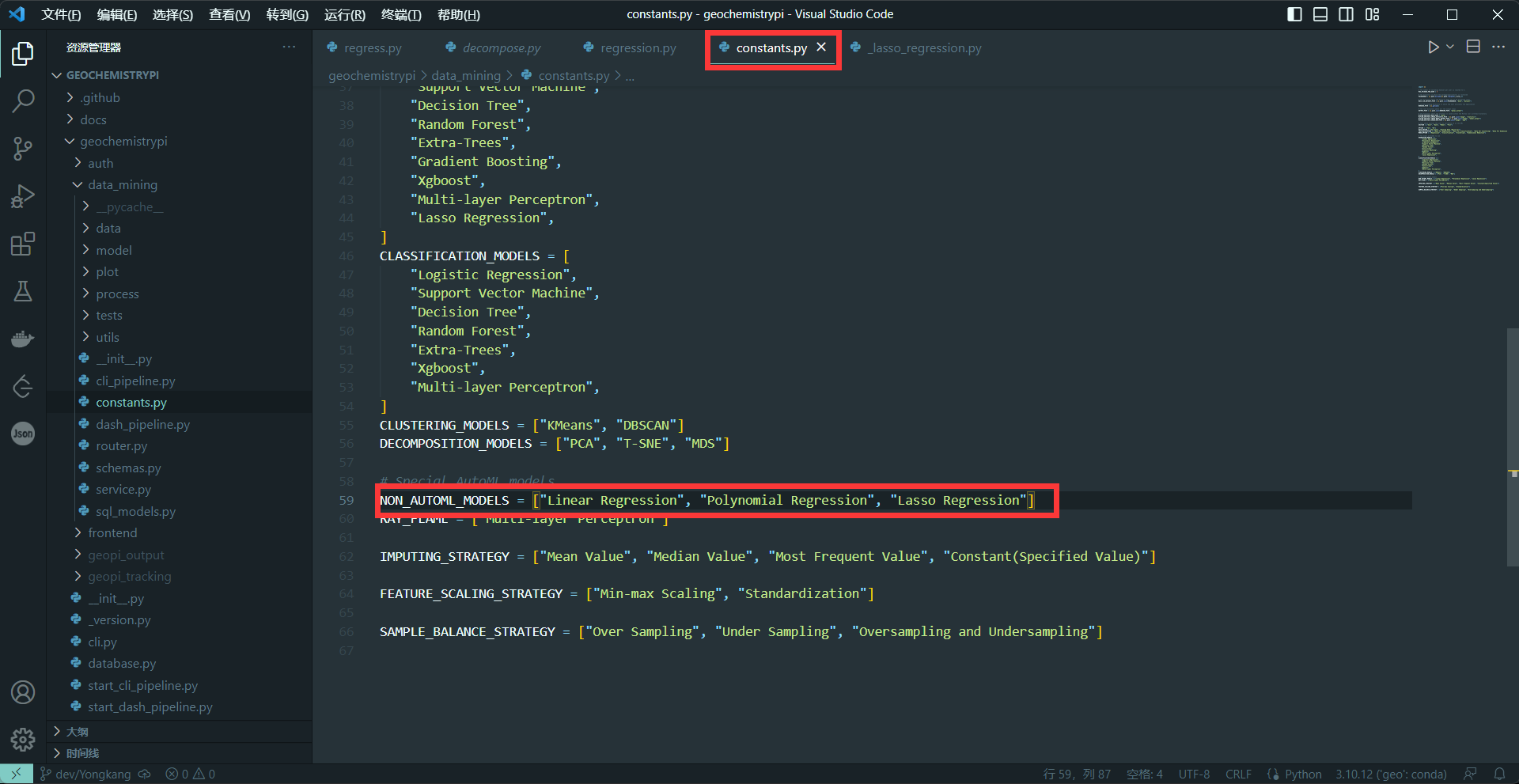
+(2)set NON_AUTOML_MODELS
+Because this is a tutorial without automatic parameters, you need to add the model name in the NON_AUTOML_MODELS.
+**eg:**
+
+
+## 3. Test model
+After the model is added, it can be tested. If the test reports an error, it needs to be checked. If there is no error, it can be submitted.
+
+## 4. Completed Pull Request
+After the model test is correct, you can complete the pull request according to the puu document instructions in [Geochemistry π](https://geochemistrypi.readthedocs.io/en/latest/index.html)
+
+
+## 5. Precautions
+**Note1:** This tutorial only discusses the general process of adding a model, and the specific addition needs to be combined with the actual situation of the model to accurately add relevant codes.
+**Note2:** If there are unclear situations and problems during the adding process, communicate with other people in time to solve them.
diff --git a/docs/source/For User/Model Example/Decomposition/decomposition.md b/docs/source/For User/Model Example/Decomposition/decomposition.md
index 01adc009..1e28db3d 100644
--- a/docs/source/For User/Model Example/Decomposition/decomposition.md
+++ b/docs/source/For User/Model Example/Decomposition/decomposition.md
@@ -431,3 +431,547 @@ By inputting different component numbers, results of PCA are obtained automatica

Figure 1 PCA Example
+
+
+
+## t-distributed Stochastic Neighbor Embedding (T-SNE)
+
+
+
+### Table of Contents
+
+- [Table of Contents](#table-of-contents)
+- [1. t-distributed Stochastic Neighbor Embedding (T-SNE)](#1-t-distributed-stochastic-neighbor-embedding-t-sne)
+- [2. Preparation](#2-preparation)
+- [3. NAN value process](#3-nan-value-process)
+- [4. Feature engineering](#4-feature-engineering)
+- [5. Model Selection](#5-model-selection)
+- [6. T-SNE](#6-t-sne)
+
+### 1. t-distributed Stochastic Neighbor Embedding (T-SNE)
+
+**t-distributed Stochastic Neighbor Embedding** is usually known as T-SNE. T-SNE is an unsupervised learning method, in which the training data we feed to the algorithm does not need the desired labels. T-SNE is a machine learning algorithm used for dimensionality reduction and visualization of high-dimensional data.
+It represents the similarity between data points in the high-dimensional space using a Gaussian distribution, creating a probability distribution by measuring this similarity. In the low-dimensional space, T-SNE reconstructs this similarity distribution using the t-distribution. T-SNE aims to preserve the local relationships between data points, ensuring that similar points in the high-dimensional space remain similar in the low-dimensional space.
+
+**Note:** This part would show the whole process of T-SNE, including data-processing and model-running.
+
+### 2. Preparation
+
+First, after ensuring the Geochemistry Pi framework has been installed successfully (if not, please see docs ), we run the python framework in command line interface to process our program: If you do not input own data, you can run:
+
+```
+geochemistrypi data-mining
+```
+
+If you prepare to input own data, you can run:
+
+```
+geochemistrypi data-mining --data your_own_data_set.xlsx
+```
+
+The command line interface would show:
+
+```
+-*-*- Built-in Training Data Option-*-*-
+1 - Data For Regression
+2 - Data For Classification
+3 - Data For Clustering
+4 - Data For Dimensional Reduction
+(User) ➜ @Number: 4
+```
+
+You have to choose **Data For Dimensional Reduction** and press **4** . The command line interface would show:
+
+```
+Successfully loading the built-in training data set 'Data_Decomposition.xlsx'.
+--------------------
+Index - Column Name
+1 - CITATION
+2 - SAMPLE NAME
+3 - Label
+4 - Notes
+5 - LATITUDE
+6 - LONGITUDE
+ ...
+45 - PB(PPM)
+46 - TH(PPM)
+47 - U(PPM)
+--------------------
+(Press Enter key to move forward.)
+```
+
+Here, we just need to press any keyboard to continue.
+
+```
+-*-*- World Map Projection -*-*-
+World Map Projection for A Specific Element Option:
+1 - Yes
+2 - No
+(Plot) ➜ @Number:
+```
+
+We can choose map projection if we need a world map projection for a specific element option. Choose yes, we can choose an element to map. Choose no, skip to the next mode. More information of the map projection can be seen in map projection. In this tutorial, we skip it and gain output as:
+
+```
+-*-*- Data Selection -*-*-
+--------------------
+Index - Column Name
+1 - CITATION
+2 - SAMPLE NAME
+3 - Label
+4 - Notes
+5 - LATITUDE
+6 - LONGITUDE
+ ...
+45 - PB(PPM)
+46 - TH(PPM)
+47 - U(PPM)
+--------------------
+Select the data range you want to process.
+Input format:
+Format 1: "[**, **]; **; [**, **]", such as "[1, 3]; 7; [10, 13]" --> you want to deal with the columns 1, 2, 3, 7, 10, 11, 12, 13
+Format 2: "xx", such as "7" --> you want to deal with the columns 7
+```
+
+Two options are offered. For T-SNE, the Format 1 method is more useful in multiple dimensional reduction. As a tutorial, we input **[10, 15]** as an example.
+**Note: [start_col_num, end_col_num]**
+
+The selected feature information would be given:
+
+```
+--------------------
+Index - Column Name
+10 - AL2O3(WT%)
+11 - CR2O3(WT%)
+12 - FEOT(WT%)
+13 - CAO(WT%)
+14 - MGO(WT%)
+15 - MNO(WT%)
+--------------------
+```
+
+```
+The Selected Data Set:
+ AL2O3(WT%) CR2O3(WT%) FEOT(WT%) CAO(WT%) MGO(WT%) MNO(WT%)
+0 3.936000 1.440 3.097000 18.546000 18.478000 0.083000
+1 3.040000 0.578 3.200000 20.235000 17.277000 0.150000
+2 7.016561 NaN 3.172049 20.092611 15.261175 0.102185
+3 3.110977 NaN 2.413834 22.083843 17.349203 0.078300
+4 6.971044 NaN 2.995074 20.530008 15.562149 0.096700
+.. ... ... ... ... ... ...
+104 2.740000 0.060 4.520000 23.530000 14.960000 0.060000
+105 5.700000 0.690 2.750000 20.120000 16.470000 0.120000
+106 0.230000 2.910 2.520000 19.700000 18.000000 0.130000
+107 2.580000 0.750 2.300000 22.100000 16.690000 0.050000
+108 6.490000 0.800 2.620000 20.560000 14.600000 0.070000
+
+[109 rows x 6 columns]
+```
+
+After continuing with any key, basic information of selected data would be shown:
+
+```
+Basic Statistical Information:
+
+RangeIndex: 109 entries, 0 to 108
+Data columns (total 6 columns):
+ # Column Non-Null Count Dtype
+--- ------ -------------- -----
+ 0 AL2O3(WT%) 109 non-null float64
+ 1 CR2O3(WT%) 98 non-null float64
+ 2 FEOT(WT%) 109 non-null float64
+ 3 CAO(WT%) 109 non-null float64
+ 4 MGO(WT%) 109 non-null float64
+ 5 MNO(WT%) 109 non-null float64
+dtypes: float64(6)
+memory usage: 5.2 KB
+None
+Some basic statistic information of the designated data set:
+ AL2O3(WT%) CR2O3(WT%) FEOT(WT%) CAO(WT%) MGO(WT%) MNO(WT%)
+count 109.000000 98.000000 109.000000 109.000000 109.000000 109.000000
+mean 4.554212 0.956426 2.962310 21.115756 16.178044 0.092087
+std 1.969756 0.553647 1.133967 1.964380 1.432886 0.054002
+min 0.230000 0.000000 1.371100 13.170000 12.170000 0.000000
+25% 3.110977 0.662500 2.350000 20.310000 15.300000 0.063075
+50% 4.720000 0.925000 2.690000 21.223500 15.920000 0.090000
+75% 6.233341 1.243656 3.330000 22.185450 16.816000 0.110000
+max 8.110000 3.869550 8.145000 25.362000 23.528382 0.400000
+Successfully calculate the pair-wise correlation coefficient among the selected columns.
+Save figure 'Correlation Plot' in dir.
+Successfully store 'Correlation Plot' in 'Correlation Plot.xlsx' in dir.
+Successfully draw the distribution plot of the selected columns.
+Save figure 'Distribution Histogram' in dir.
+Successfully store 'Distribution Histogram' in 'Distribution Histogram.xlsx' in dir.
+Successfully draw the distribution plot after log transformation of the selected columns.
+Save figure 'Distribution Histogram After Log Transformation' in dir.
+Successfully store 'Distribution Histogram After Log Transformation' in 'Distribution Histogram After Log Transformation.xlsx' in dir.
+Successfully store 'Data Original' in 'Data Original.xlsx' in dir.
+Successfully store 'Data Selected' in 'Data Selected.xlsx' in dir.
+```
+
+### 3. NAN value process
+
+Check the NAN values would be helpful for later analysis. In Geochemistry π frame, this option is finished automatically.
+
+```
+-*-*- Imputation -*-*-
+Check which column has null values:
+--------------------
+AL2O3(WT%) False
+CR2O3(WT%) True
+FEOT(WT%) False
+CAO(WT%) False
+MGO(WT%) False
+MNO(WT%) False
+dtype: bool
+--------------------
+The ratio of the null values in each column:
+--------------------
+CR2O3(WT%) 0.100917
+AL2O3(WT%) 0.000000
+FEOT(WT%) 0.000000
+CAO(WT%) 0.000000
+MGO(WT%) 0.000000
+MNO(WT%) 0.000000
+dtype: float64
+--------------------
+Note: you'd better use imputation techniques to deal with the missing values.
+```
+
+Several strategies are offered for processing the missing values, including:
+
+```
+-*-*- Strategy for Missing Values -*-*-
+1 - Mean Value
+2 - Median Value
+3 - Most Frequent Value
+4 - Constant(Specified Value)
+Which strategy do you want to apply?
+(Data) ➜ @Number:1
+```
+
+We choose the mean Value in this example and the input data be processed automatically as:
+
+```
+Successfully fill the missing values with the mean value of each feature column respectively.
+(Press Enter key to move forward.)
+```
+
+```python
+-*-*- Hypothesis Testing on Imputation Method -*-*-
+Null Hypothesis: The distributions of the data set before and after imputing remain the same.
+Thoughts: Check which column rejects null hypothesis.
+Statistics Test Method: Kruskal Test
+Significance Level: 0.05
+The number of iterations of Monte Carlo simulation: 100
+The size of the sample for each iteration (half of the whole data set): 54
+Average p-value:
+AL2O3(WT%) 1.0
+CR2O3(WT%) 0.9327453056346102
+FEOT(WT%) 1.0
+CAO(WT%) 1.0
+MGO(WT%) 1.0
+MNO(WT%) 1.0
+Note: 'p-value < 0.05' means imputation method doesn't apply to that column.
+The columns which rejects null hypothesis: None
+Successfully draw the respective probability plot (origin vs. impute) of the selected columns
+Save figure 'Probability Plot' in dir.
+Successfully store 'Probability Plot' in 'Probability Plot.xlsx' in dir.
+
+RangeIndex: 109 entries, 0 to 108
+Data columns (total 6 columns):
+ # Column Non-Null Count Dtype
+--- ------ -------------- -----
+ 0 AL2O3(WT%) 109 non-null float64
+ 1 CR2O3(WT%) 109 non-null float64
+ 2 FEOT(WT%) 109 non-null float64
+ 3 CAO(WT%) 109 non-null float64
+ 4 MGO(WT%) 109 non-null float64
+ 5 MNO(WT%) 109 non-null float64
+dtypes: float64(6)
+memory usage: 5.2 KB
+None
+Some basic statistic information of the designated data set:
+ AL2O3(WT%) CR2O3(WT%) FEOT(WT%) CAO(WT%) MGO(WT%) MNO(WT%)
+count 109.000000 109.000000 109.000000 109.000000 109.000000 109.000000
+mean 4.554212 0.956426 2.962310 21.115756 16.178044 0.092087
+std 1.969756 0.524695 1.133967 1.964380 1.432886 0.054002
+min 0.230000 0.000000 1.371100 13.170000 12.170000 0.000000
+25% 3.110977 0.680000 2.350000 20.310000 15.300000 0.063075
+50% 4.720000 0.956426 2.690000 21.223500 15.920000 0.090000
+75% 6.233341 1.170000 3.330000 22.185450 16.816000 0.110000
+max 8.110000 3.869550 8.145000 25.362000 23.528382 0.400000
+Successfully store 'Data Selected Imputed' in 'Data Selected Imputed.xlsx' in dir.
+```
+
+### 4. Feature engineering
+
+The next step is the feature engineering options.
+
+```python
+-*-*- Feature Engineering -*-*-
+The Selected Data Set:
+--------------------
+Index - Column Name
+1 - AL2O3(WT%)
+2 - CR2O3(WT%)
+3 - FEOT(WT%)
+4 - CAO(WT%)
+5 - MGO(WT%)
+6 - MNO(WT%)
+--------------------
+Feature Engineering Option:
+1 - Yes
+2 - No
+(Data) ➜ @Number: 1
+```
+
+Feature engineering options are essential for data analysis. We choose Yes and naming new features:
+
+```python
+Selected data set:
+a - AL2O3(WT%)
+b - CR2O3(WT%)
+c - FEOT(WT%)
+d - CAO(WT%)
+e - MGO(WT%)
+f - MNO(WT%)
+Name the constructed feature (column name), like 'NEW-COMPOUND':
+@input: new
+```
+
+Considering actual need for constructing several new geochemical indexes. We can set up some new indexes. Here, we would set up a new index by AL2O3/CAO via keyboard options with a/d.
+
+```python
+-*-*- Feature Engineering -*-*-
+The Selected Data Set:
+--------------------
+Index - Column Name
+1 - AL2O3(WT%)
+2 - CR2O3(WT%)
+3 - FEOT(WT%)
+4 - CAO(WT%)
+5 - MGO(WT%)
+6 - MNO(WT%)
+--------------------
+Feature Engineering Option:
+1 - Yes
+2 - No
+(Data) ➜ @Number: 1
+Selected data set:
+a - AL2O3(WT%)
+b - CR2O3(WT%)
+c - FEOT(WT%)
+d - CAO(WT%)
+e - MGO(WT%)
+f - MNO(WT%)
+Name the constructed feature (column name), like 'NEW-COMPOUND':
+@input: new
+Build up new feature with the combination of basic arithmatic operators, including '+', '-', '*', '/', '()'.
+Input example 1: a * b - c
+--> Step 1: Multiply a column with b column;
+--> Step 2: Subtract c from the result of Step 1;
+Input example 2: (d + 5 * f) / g
+--> Step 1: Multiply 5 with f;
+--> Step 2: Plus d column with the result of Step 1;
+--> Step 3: Divide the result of Step 1 by g;
+Input example 3: pow(a, b) + c * d
+--> Step 1: Raise the base a to the power of the exponent b;
+--> Step 2: Multiply the value of c by the value of d;
+--> Step 3: Add the result of Step 1 to the result of Step 2;
+Input example 4: log(a)/b - c
+--> Step 1: Take the logarithm of the value a;
+--> Step 2: Divide the result of Step 1 by the value of b;
+--> Step 3: Subtract the value of c from the result of Step 2;
+You can use mean(x) to calculate the average value.
+@input: a/d
+```
+
+```python
+Successfully construct a new feature new.
+0 0.212229
+1 0.150235
+2 0.349211
+3 0.140871
+4 0.339554
+ ...
+104 0.116447
+105 0.283300
+106 0.011675
+107 0.116742
+108 0.315661
+Name: new, Length: 109, dtype: float64
+```
+
+Basic information of selected data would be shown:
+
+```python
+
+RangeIndex: 109 entries, 0 to 108
+Data columns (total 7 columns):
+ # Column Non-Null Count Dtype
+--- ------ -------------- -----
+ 0 AL2O3(WT%) 109 non-null float64
+ 1 CR2O3(WT%) 109 non-null float64
+ 2 FEOT(WT%) 109 non-null float64
+ 3 CAO(WT%) 109 non-null float64
+ 4 MGO(WT%) 109 non-null float64
+ 5 MNO(WT%) 109 non-null float64
+ 6 new 109 non-null float64
+dtypes: float64(7)
+memory usage: 6.1 KB
+None
+Some basic statistic information of the designated data set:
+ AL2O3(WT%) CR2O3(WT%) FEOT(WT%) CAO(WT%) MGO(WT%) MNO(WT%) new
+count 109.000000 109.000000 109.000000 109.000000 109.000000 109.000000 109.000000
+mean 4.554212 0.956426 2.962310 21.115756 16.178044 0.092087 0.219990
+std 1.969756 0.524695 1.133967 1.964380 1.432886 0.054002 0.101476
+min 0.230000 0.000000 1.371100 13.170000 12.170000 0.000000 0.011675
+25% 3.110977 0.680000 2.350000 20.310000 15.300000 0.063075 0.148707
+50% 4.720000 0.956426 2.690000 21.223500 15.920000 0.090000 0.218100
+75% 6.233341 1.170000 3.330000 22.185450 16.816000 0.110000 0.306383
+max 8.110000 3.869550 8.145000 25.362000 23.528382 0.400000 0.407216
+```
+
+Do not continue to establish new features:
+
+```python
+Do you want to continue to build a new feature?
+1 - Yes
+2 - No
+(Data) ➜ @Number: 2
+Successfully store 'Data Selected Imputed Feature-Engineering' in 'Data Selected Imputed Feature-Engineering.xlsx' in dir.
+Exit Feature Engineering Mode.
+```
+
+### 5. Model Selection
+
+Select dimensionality reduction
+
+```python
+-*-*- Mode Selection -*-*-
+1 - Regression
+2 - Classification
+3 - Clustering
+4 - Dimensional Reduction
+(Model) ➜ @Number: 4
+```
+
+Scaling features on set X.In this tutorial, we skip it
+
+```python
+-*-*- Feature Scaling on X Set -*-*-
+1 - Yes
+2 - No
+(Data) ➜ @Number: 1
+```
+
+```python
+-*-*- Which strategy do you want to apply?-*-*-
+1 - Min-max Scaling
+2 - Standardization
+(Data) ➜ @Number: 2
+```
+
+```python
+Data Set After Scaling:
+ AL2O3(WT%) CR2O3(WT%) FEOT(WT%) CAO(WT%) MGO(WT%) MNO(WT%)
+0 -0.315302 0.925885 0.119326 -1.314219 1.612536 -0.169053
+1 -0.772282 -0.724562 0.210577 -0.450434 0.770496 1.077372
+2 1.255852 0.000000 0.185815 -0.523255 -0.642832 0.187847
+3 -0.736082 0.000000 -0.485913 0.495097 0.821118 -0.256489
+4 1.232638 0.000000 0.029026 -0.299562 -0.431813 0.085813
+.. ... ... ... ... ... ...
+104 -0.925288 -1.716362 1.380010 1.234688 -0.853990 -0.596931
+105 0.584377 -0.510119 -0.188093 -0.509247 0.204695 0.519271
+106 -2.205444 3.740453 -0.391857 -0.724043 1.277403 0.705305
+107 -1.006892 -0.395238 -0.586763 0.503360 0.358941 -0.782964
+108 0.987295 -0.299505 -0.303264 -0.284223 -1.106392 -0.410897
+
+[109 rows x 6 columns]
+Basic Statistical Information:
+Some basic statistic information of the designated data set:
+ AL2O3(WT%) CR2O3(WT%) FEOT(WT%) CAO(WT%) MGO(WT%) MNO(WT%)
+count 1.090000e+02 1.090000e+02 1.090000e+02 1.090000e+02 1.090000e+02 1.090000e+02
+mean 1.415789e-16 -8.912341e-17 4.216810e-16 4.685345e-17 -6.722451e-17 -1.874138e-16
+std 1.004619e+00 1.004619e+00 1.004619e+00 1.004619e+00 1.004619e+00 1.004619e+00
+min -2.205444e+00 -1.831243e+00 -1.409706e+00 -4.063601e+00 -2.810103e+00 -1.713133e+00
+25% -7.360817e-01 -5.292655e-01 -5.424660e-01 -4.120778e-01 -6.156105e-01 -5.397252e-01
+50% 8.455546e-02 0.000000e+00 -2.412486e-01 5.510241e-02 -1.809186e-01 -3.882964e-02
+75% 8.563927e-01 4.089238e-01 3.257487e-01 5.470608e-01 4.472813e-01 3.332377e-01
+max 1.813530e+00 5.577677e+00 4.591518e+00 2.171605e+00 5.153439e+00 5.728214e+00
+Successfully store 'X With Scaling' in 'X With Scaling.xlsx' in dir.
+```
+
+
+
+### 6. T-SNE
+
+Select T-SNE.
+
+```python
+-*-*- Model Selection -*-*-:
+1 - PCA
+2 - T-SNE
+3 - MDS
+4 - All models above to be trained
+Which model do you want to apply?(Enter the Corresponding Number)
+(Model) ➜ @Number: 2
+```
+
+Input the Hyper-parameters.
+
+```python
+-*-*- T-SNE - Hyper-parameters Specification -*-*-
+N Components: This parameter specifies the number of components to retain after dimensionality reduction.
+Please specify the number of components to retain. A good starting range could be between 2 and 10, such as 4.
+(Model) ➜ N Components: 4
+Perplexity: This parameter is related to the number of nearest neighbors that each point considers when computing the probabilities.
+Please specify the perplexity. A good starting range could be between 5 and 50, such as 30.
+(Model) ➜ Perplexity: 30
+Learning Rate: This parameter controls the step size during the optimization process.
+Please specify the learning rate. A good starting range could be between 10 and 1000, such as 200.
+(Model) ➜ Learning Rate: 200
+Number of Iterations: This parameter controls how many iterations the optimization will run for.
+Please specify the number of iterations. A good starting range could be between 250 and 1000, such as 500.
+(Model) ➜ Number of Iterations: 500
+Early Exaggeration: This parameter controls how tight natural clusters in the original space are in the embedded space and how much space will be between them.
+Please specify the early exaggeration. A good starting range could be between 5 and 50, such as 12.
+(Model) ➜ Early Exaggeration: 12
+```
+
+Running this model.
+
+```python
+*-**-* T-SNE is running ... *-**-*
+Expected Functionality:
++ Model Persistence
+Successfully store 'Hyper Parameters - T-SNE' in 'Hyper Parameters - T-SNE.txt' in dir.
+-----* Reduced Data *-----
+ Dimension 1 Dimension 2 Dimension 3 Dimension 4
+0 -10.623264 -17.686243 51.568302 1.087756
+1 21.165867 50.837616 -2.456909 69.025017
+2 35.604347 -36.303295 -11.843322 -21.651896
+3 -25.802412 28.442354 44.452190 18.172804
+4 -23.921820 -48.037205 -13.831066 14.313259
+.. ... ... ... ...
+104 43.789333 -8.022134 26.877687 -7.914544
+105 -12.640723 14.591939 -43.875713 4.952276
+106 359.025940 -895.016479 -461.668243 491.801666
+107 -7.346601 35.262451 25.139845 1.618280
+108 188.163788 66.346474 -9.461174 190.716721
+
+[109 rows x 4 columns]
+Successfully store 'X Reduced' in 'X Reduced.xlsx' in dir.
+-----* Model Persistence *-----
+Successfully store 'T-SNE' in 'T-SNE.pkl' in dir.
+Successfully store 'T-SNE' in 'T-SNE.joblib' in dir.
+```
+
+```python
+-*-*- Transform Pipeline -*-*-
+Build the transform pipeline according to the previous operations.
+Successfully store 'Transform Pipeline Configuration' in dir.
+Successfully store 'Transform Pipeline' in 'Transform Pipeline.pkl' in dir.
+Successfully store 'Transform Pipeline' in 'Transform Pipeline.joblib' in dir.
+```
diff --git a/docs/source/conf.py b/docs/source/conf.py
index c5382f81..b8e5e3f5 100644
--- a/docs/source/conf.py
+++ b/docs/source/conf.py
@@ -31,8 +31,9 @@
# https://www.sphinx-doc.org/en/master/usage/configuration.html#options-for-html-output
-html_theme = "furo"
+# html_theme = "furo"
# html_theme = 'classic'
+html_theme = "sphinx_rtd_theme"
html_theme_path = [sphinx_rtd_theme.get_html_theme_path()]
html_static_path = ["_static"]
@@ -59,11 +60,11 @@
# autodoc_mock_imports = ["geochemistrypi"]
sys.path.insert(0, os.path.abspath("../.."))
# sys.path.insert(0, os.path.abspath(project_path))
-print(os.path.abspath(project_path))
-sys.path.insert(1, os.path.abspath("../../geochemistrypi/"))
-sys.path.insert(2, os.path.abspath("../../geochemistrypi/data_mining/"))
-sys.path.insert(3, os.path.abspath(".."))
-sys.path.append("../geochemistrypy/geochemistrypy")
+# print(os.path.abspath(project_path))
+# sys.path.insert(1, os.path.abspath("../../geochemistrypi/"))
+# sys.path.insert(2, os.path.abspath("../../geochemistrypi/data_mining/"))
+# sys.path.insert(3, os.path.abspath(".."))
+# sys.path.append("../geochemistrypy/geochemistrypy")
# ...
apidoc_module_dir = project_path
diff --git a/docs/source/index.rst b/docs/source/index.rst
index b62f4afd..3954b500 100644
--- a/docs/source/index.rst
+++ b/docs/source/index.rst
@@ -23,7 +23,6 @@ Welcome to Geochemistry π
Model Example
Contact Us
Docs Link
- API
.. toctree::
:maxdepth: 3
@@ -35,6 +34,7 @@ Welcome to Geochemistry π
Docker Deployment
Algorithm Functionality Addition
Complete Pull Request
+ Add New Model To Framework
Docs Link
Indices and tables
diff --git a/docs/source/python_apis/geochemistrypi.data_mining.data.rst b/docs/source/python_apis/geochemistrypi.data_mining.data.rst
index 5bd2c2df..0474173a 100644
--- a/docs/source/python_apis/geochemistrypi.data_mining.data.rst
+++ b/docs/source/python_apis/geochemistrypi.data_mining.data.rst
@@ -28,6 +28,14 @@ geochemistrypi.data\_mining.data.imputation module
:undoc-members:
:show-inheritance:
+geochemistrypi.data\_mining.data.inference module
+-------------------------------------------------
+
+.. automodule:: geochemistrypi.data_mining.data.inference
+ :members:
+ :undoc-members:
+ :show-inheritance:
+
geochemistrypi.data\_mining.data.preprocessing module
-----------------------------------------------------
From 967ba167133a2623088e1c39a9df74e4c647c3b2 Mon Sep 17 00:00:00 2001
From: jin <571979568@qq.com>
Date: Tue, 26 Dec 2023 18:21:30 +0800
Subject: [PATCH 2/4] feat : Add BayesianRidgeRegression Algorithm to Project
Framework
---
geochemistrypi/data_mining/constants.py | 1 +
.../_bayesianridge_regression.py | 66 +++++++
.../data_mining/model/regression.py | 172 +++++++++++++++++-
geochemistrypi/data_mining/process/regress.py | 18 ++
4 files changed, 256 insertions(+), 1 deletion(-)
create mode 100644 geochemistrypi/data_mining/model/func/algo_regression/_bayesianridge_regression.py
diff --git a/geochemistrypi/data_mining/constants.py b/geochemistrypi/data_mining/constants.py
index 6822085a..2b8d1fc6 100644
--- a/geochemistrypi/data_mining/constants.py
+++ b/geochemistrypi/data_mining/constants.py
@@ -44,6 +44,7 @@
"Lasso Regression",
"Elastic Net",
"SGD Regression",
+ "BayesianRidge Regression",
# "Bagging Regression",
# "Decision Tree",
# Histogram-based Gradient Boosting,
diff --git a/geochemistrypi/data_mining/model/func/algo_regression/_bayesianridge_regression.py b/geochemistrypi/data_mining/model/func/algo_regression/_bayesianridge_regression.py
new file mode 100644
index 00000000..42967319
--- /dev/null
+++ b/geochemistrypi/data_mining/model/func/algo_regression/_bayesianridge_regression.py
@@ -0,0 +1,66 @@
+from typing import Dict
+
+from rich import print
+
+from ....constants import SECTION
+from ....data.data_readiness import bool_input, float_input
+
+
+def bayesian_ridge_manual_hyper_parameters() -> Dict:
+ """Manually set hyperparameters for Bayesian Ridge Regression.
+
+ Returns
+ -------
+ hyper_parameters : dict
+ The hyperparameters.
+ """
+ # print("Maximum Number of Iterations: The maximum number of iterations for training. The default is 300.")
+ # max_iter = num_input(SECTION[2], "@Max Iterations: ")
+
+ print("Tolerance: The tolerance for stopping criteria. A good starting value could be between 1e-6 and 1e-3, such as 1e-4. The default is 0.001.")
+ tol = float_input(1e-4, SECTION[2], "@Tol: ")
+
+ print("Alpha 1: Hyperparameter for the Gamma distribution prior over the alpha parameter. A good starting value could be between 1e-6 and 1e-3, such as 1e-6. The default is 0.000001.")
+ alpha_1 = float_input(1e-6, SECTION[2], "@Alpha 1: ")
+
+ print("Alpha 2: Hyperparameter for the Gamma distribution prior over the beta parameter. A good starting value could be between 1e-6 and 1e-3, such as 1e-6. The default is 0.000001.")
+ alpha_2 = float_input(1e-6, SECTION[2], "@Alpha 2: ")
+
+ print("Lambda 1: Hyperparameter for the Gaussian distribution prior over the weights. A good starting value could be between 1e-6 and 1e-3, such as 1e-6. The default is 0.000001.")
+ lambda_1 = float_input(1e-6, SECTION[2], "@Lambda 1: ")
+
+ print("Lambda 2: Hyperparameter for the Gamma distribution prior over the noise. A good starting value could be between 1e-6 and 1e-3, such as 1e-6. The default is 0.000001.")
+ lambda_2 = float_input(1e-6, SECTION[2], "@Lambda 2: ")
+
+ print("Alpha Init: Initial guess for alpha. The default is 0.000001.")
+ alpha_init = float_input(1.0, SECTION[2], "@Alpha Init: ")
+
+ print("Lambda Init: Initial guess for lambda. The default is 0.000001.")
+ lambda_init = float_input(1.0, SECTION[2], "@Lambda Init: ")
+
+ print("Compute Score: Whether to compute the objective function at each step. The default is False.")
+ compute_score = bool_input(SECTION[2])
+
+ print("Fit Intercept: Whether to fit an intercept term. It is generally recommended to set it as True.")
+ fit_intercept = bool_input(SECTION[2])
+
+ print("Copy X: Whether to copy X in fit. The default is True.")
+ copy_X = bool_input(SECTION[2])
+
+ print("Verbose: Verbosity of the solution. The default is False.")
+ verbose = bool_input(SECTION[2])
+
+ hyper_parameters = {
+ "tol": tol,
+ "alpha_1": alpha_1,
+ "alpha_2": alpha_2,
+ "lambda_1": lambda_1,
+ "lambda_2": lambda_2,
+ "alpha_init": alpha_init,
+ "lambda_init": lambda_init,
+ "compute_score": compute_score,
+ "fit_intercept": fit_intercept,
+ "copy_X": copy_X,
+ "verbose": verbose,
+ }
+ return hyper_parameters
diff --git a/geochemistrypi/data_mining/model/regression.py b/geochemistrypi/data_mining/model/regression.py
index 834347b6..b17aacae 100644
--- a/geochemistrypi/data_mining/model/regression.py
+++ b/geochemistrypi/data_mining/model/regression.py
@@ -11,7 +11,7 @@
from multipledispatch import dispatch
from rich import print
from sklearn.ensemble import ExtraTreesRegressor, GradientBoostingRegressor, RandomForestRegressor
-from sklearn.linear_model import ElasticNet, Lasso, LinearRegression, SGDRegressor
+from sklearn.linear_model import BayesianRidge, ElasticNet, Lasso, LinearRegression, SGDRegressor
from sklearn.neighbors import KNeighborsRegressor
from sklearn.neural_network import MLPRegressor
from sklearn.preprocessing import PolynomialFeatures
@@ -21,6 +21,7 @@
from ..constants import MLFLOW_ARTIFACT_IMAGE_MODEL_OUTPUT_PATH, RAY_FLAML
from ..utils.base import clear_output, save_data, save_fig, save_text
from ._base import LinearWorkflowMixin, TreeWorkflowMixin, WorkflowBase
+from .func.algo_regression._bayesianridge_regression import bayesian_ridge_manual_hyper_parameters
from .func.algo_regression._common import cross_validation, plot_predicted_vs_actual, plot_residuals, score
from .func.algo_regression._decision_tree import decision_tree_manual_hyper_parameters
from .func.algo_regression._elastic_net import elastic_net_manual_hyper_parameters
@@ -3740,3 +3741,172 @@ def special_components(self, is_automl: bool, **kwargs) -> None:
)
else:
pass
+
+
+class BayesianRidgeRegression(RegressionWorkflowBase):
+ """The automation workflow of using Bayesian ridge regression algorithm to make insightful products."""
+
+ name = "BayesianRidge Regression"
+ special_function = []
+
+ def __init__(
+ self,
+ tol: float = 0.001,
+ alpha_1: float = 0.000001,
+ alpha_2: float = 0.000001,
+ lambda_1: float = 0.000001,
+ lambda_2: float = 0.000001,
+ alpha_init: float = None,
+ lambda_init: float = None,
+ compute_score: bool = False,
+ fit_intercept: bool = True,
+ copy_X: bool = True,
+ verbose: bool = False,
+ ) -> None:
+ """
+ Parameters
+ ------------
+ max_iter : int, default=None
+ Maximum number of iterations over the complete dataset before stopping independently of any early stopping criterion. If , it corresponds to .Nonemax_iter=300
+
+ Changed in version 1.3.
+
+ tol : float, default=1e-3
+ Stop the algorithm if w has converged.
+
+ alpha_1 : float, default=1e-6
+ Hyper-parameter : shape parameter for the Gamma distribution prior over the alpha parameter.
+
+ alpha_2 : float, default=1e-6
+ Hyper-parameter : inverse scale parameter (rate parameter) for the Gamma distribution prior over the alpha parameter.
+
+ lambda_1 : float, default=1e-6
+ Hyper-parameter : shape parameter for the Gamma distribution prior over the lambda parameter.
+
+ lambda_2 : float, default=1e-6
+ Hyper-parameter : inverse scale parameter (rate parameter) for the Gamma distribution prior over the lambda parameter.
+
+ alpha_init : float, default=None
+ Initial value for alpha (precision of the noise). If not set, alpha_init is 1/Var(y).
+
+ New in version 0.22.
+
+ lambda_init : float, default=None
+ Initial value for lambda (precision of the weights). If not set, lambda_init is 1.
+
+ New in version 0.22.
+
+ compute_score : bool, default=False
+ If True, compute the log marginal likelihood at each iteration of the optimization.
+
+ fit_intercept : bool, default=True
+ Whether to calculate the intercept for this model. The intercept is not treated as a probabilistic parameter and
+ thus has no associated variance. If set to False, no intercept will be used in calculations (i.e. data is expect
+ ed to be centered).
+
+ copy_X : bool, default=True
+ If True, X will be copied; else, it may be overwritten.
+
+ verbose : bool, default=False
+ Verbose mode when fitting the model.
+
+ n_iter : int
+ Maximum number of iterations. Should be greater than or equal to 1.
+
+ References
+ ----------
+ Scikit-learn API: sklearn.linear_model.BayesianRidge
+ https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.BayesianRidge.html
+ """
+ super().__init__()
+ self.tol = tol
+ self.alpha_1 = alpha_1
+ self.alpha_2 = alpha_2
+ self.lambda_1 = lambda_1
+ self.lambda_2 = lambda_2
+ self.alpha_init = alpha_init
+ self.lambda_init = lambda_init
+ self.compute_score = compute_score
+ self.fit_intercept = fit_intercept
+ self.copy_X = copy_X
+ self.verbose = verbose
+
+ self.model = BayesianRidge(
+ tol=self.tol,
+ alpha_1=self.alpha_1,
+ alpha_2=self.alpha_2,
+ lambda_1=self.lambda_1,
+ lambda_2=self.lambda_2,
+ alpha_init=self.alpha_init,
+ lambda_init=self.lambda_init,
+ compute_score=self.compute_score,
+ fit_intercept=self.compute_score,
+ copy_X=self.copy_X,
+ verbose=self.verbose,
+ )
+
+ self.naming = BayesianRidgeRegression.name
+ self.customized = True
+ self.customized_name = "BayesianRidge Regression"
+
+ @property
+ def settings(self) -> Dict:
+ """The configuration of BayesianRidge to implement AutoML by FLAML framework."""
+ configuration = {
+ "time_budget": 10, # total running time in seconds
+ "metric": "r2",
+ "estimator_list": [self.customized_name], # list of ML learners
+ "task": "regression", # task type
+ # "log_file_name": f'{self.naming} - automl.log', # flaml log file
+ # "log_training_metric": True, # whether to log training metric
+ }
+ return configuration
+
+ @property
+ def customization(self) -> object:
+ """The customized BayesianRidge of FLAML framework."""
+ from flaml import tune
+ from flaml.data import REGRESSION
+ from flaml.model import SKLearnEstimator
+ from sklearn.linear_model import BayesianRidge
+
+ class MyBayesianRidgeRegression(SKLearnEstimator):
+ def __init__(self, task="regression", n_jobs=None, **config):
+ super().__init__(task, **config)
+ if task in REGRESSION:
+ self.estimator_class = BayesianRidge
+
+ @classmethod
+ def search_space(cls, data_size, task):
+ space = {
+ "tol": {"domain": tune.loguniform(lower=0.000001, upper=0.001), "init_value": 0.001},
+ "alpha_1": {"domain": tune.loguniform(lower=0.0001, upper=1), "init_value": 0.000001},
+ "alpha_2": {"domain": tune.loguniform(lower=0.0001, upper=1), "init_value": 0.000001},
+ "lambda_1": {"domain": tune.loguniform(lower=0.0001, upper=1), "init_value": 0.000001},
+ "lambda_2": {"domain": tune.loguniform(lower=0.0001, upper=1), "init_value": 0.000001},
+ "alpha_init": {"domain": tune.loguniform(lower=0.0001, upper=1), "init_value": 0.000001},
+ "lambda_init": {"domain": tune.loguniform(lower=0.0001, upper=1), "init_value": 0.000001},
+ "compute_score": {"domain": tune.choice([True, False]), "init_value": False},
+ "fit_intercept": {"domain": tune.choice([True, False]), "init_value": True},
+ "copy_X": {"domain": tune.choice([True, False]), "init_value": True},
+ "verbose": {"domain": tune.choice([True, False]), "init_value": False},
+ }
+ return space
+
+ return MyBayesianRidgeRegression
+
+ @classmethod
+ def manual_hyper_parameters(cls) -> Dict:
+ """Manual hyper-parameters specification."""
+ print(f"-*-*- {cls.name} - Hyper-parameters Specification -*-*-")
+ hyper_parameters = bayesian_ridge_manual_hyper_parameters()
+ clear_output()
+ return hyper_parameters
+
+ @dispatch()
+ def special_components(self, **kwargs) -> None:
+ pass
+
+ @dispatch(bool)
+ def special_components(self, is_automl: bool, **kwargs) -> None:
+ pass
diff --git a/geochemistrypi/data_mining/process/regress.py b/geochemistrypi/data_mining/process/regress.py
index 7c63ea72..d6b9c07d 100644
--- a/geochemistrypi/data_mining/process/regress.py
+++ b/geochemistrypi/data_mining/process/regress.py
@@ -8,6 +8,7 @@
from ..constants import MLFLOW_ARTIFACT_DATA_PATH, SECTION
from ..data.data_readiness import num_input
from ..model.regression import (
+ BayesianRidgeRegression,
ClassicalLinearRegression,
DecisionTreeRegression,
ElasticNetRegression,
@@ -181,6 +182,21 @@ def activate(
eta0=hyper_parameters["eta0"],
power_t=hyper_parameters["power_t"],
)
+ elif self.model_name == "BayesianRidge Regression":
+ hyper_parameters = BayesianRidgeRegression.manual_hyper_parameters()
+ self.reg_workflow = BayesianRidgeRegression(
+ tol=hyper_parameters["tol"],
+ alpha_1=hyper_parameters["alpha_1"],
+ alpha_2=hyper_parameters["alpha_2"],
+ lambda_1=hyper_parameters["lambda_1"],
+ lambda_2=hyper_parameters["lambda_2"],
+ alpha_init=hyper_parameters["alpha_init"],
+ lambda_init=hyper_parameters["lambda_init"],
+ compute_score=hyper_parameters["compute_score"],
+ fit_intercept=hyper_parameters["fit_intercept"],
+ copy_X=hyper_parameters["copy_X"],
+ verbose=hyper_parameters["verbose"],
+ )
self.reg_workflow.show_info()
@@ -252,6 +268,8 @@ def activate(
self.reg_workflow = ElasticNetRegression()
elif self.model_name == "SGD Regression":
self.reg_workflow = SGDRegression()
+ elif self.model_name == "BayesianRidge Regression":
+ self.reg_workflow = BayesianRidgeRegression()
self.reg_workflow.show_info()
From d1acbcc8e237b8add97da4271e42fa5eb862c129 Mon Sep 17 00:00:00 2001
From: jin <571979568@qq.com>
Date: Thu, 28 Dec 2023 19:06:12 +0800
Subject: [PATCH 3/4] docs: Adjust the style of the document
---
docs/source/conf.py | 4 ++--
1 file changed, 2 insertions(+), 2 deletions(-)
diff --git a/docs/source/conf.py b/docs/source/conf.py
index b8e5e3f5..644810b1 100644
--- a/docs/source/conf.py
+++ b/docs/source/conf.py
@@ -31,9 +31,9 @@
# https://www.sphinx-doc.org/en/master/usage/configuration.html#options-for-html-output
-# html_theme = "furo"
+html_theme = "furo"
# html_theme = 'classic'
-html_theme = "sphinx_rtd_theme"
+# html_theme = "sphinx_rtd_theme"
html_theme_path = [sphinx_rtd_theme.get_html_theme_path()]
html_static_path = ["_static"]
From e9fc215419e9b164c5aa870d92404cc18b697a6d Mon Sep 17 00:00:00 2001
From: jin <571979568@qq.com>
Date: Thu, 28 Dec 2023 19:37:31 +0800
Subject: [PATCH 4/4] docs: Delete the Algorithm Functionality Addition
document and modify the content of the dimensionality reduction algorithm
module
---
.../data_mining_geochemistrypi_020.md | 2 -
.../Decomposition/decomposition.md | 437 +-----------------
docs/source/index.rst | 1 -
3 files changed, 2 insertions(+), 438 deletions(-)
delete mode 100644 docs/source/For Developer/Algorithm Functionality Addition Procedure_geochemistrypi 0.2.0_CN/data_mining_geochemistrypi_020.md
diff --git a/docs/source/For Developer/Algorithm Functionality Addition Procedure_geochemistrypi 0.2.0_CN/data_mining_geochemistrypi_020.md b/docs/source/For Developer/Algorithm Functionality Addition Procedure_geochemistrypi 0.2.0_CN/data_mining_geochemistrypi_020.md
deleted file mode 100644
index 8a89e282..00000000
--- a/docs/source/For Developer/Algorithm Functionality Addition Procedure_geochemistrypi 0.2.0_CN/data_mining_geochemistrypi_020.md
+++ /dev/null
@@ -1,2 +0,0 @@
-Data-mining v0.2.0 (Updating)
-==========
diff --git a/docs/source/For User/Model Example/Decomposition/decomposition.md b/docs/source/For User/Model Example/Decomposition/decomposition.md
index 1e28db3d..c03afc02 100644
--- a/docs/source/For User/Model Example/Decomposition/decomposition.md
+++ b/docs/source/For User/Model Example/Decomposition/decomposition.md
@@ -1,440 +1,7 @@
# Decomposition
-## Principal Component Analysis (PCA)
-Principal component analysis is usually known as PCA. PCA is an unsupervised learning method, in which the training data we feed to the algorithm does not need the desired labels. The aim of PCA is to reduce the dimension of high-dimensional input data. For example, there are x1, x2, x3, x4 and y, up to four columns of data. However, not all kinds of data are essential for the label y. So the PCA could be useful to abandon less important x for regressing/classifying y and accelerate data analysis.
-
-
Note : This part would show the whole process of PCA, including data-processing and model-running.
-
-## Preparation
-
-First, after ensuring the Geochemistry Pi framework has been installed successfully (if not, please see docs ), we run the python framework in command line interface to process our program:
-If you do not input own data, you can run
-
-```bash
-geochemistrypi data-mining
-```
-
-If you prepare to input own data, you can run
-
-```bash
-geochemistrypi data-mining --data your_own_data_set.xlsx
-```
-
-The command line interface would show
-
-```bash
--*-*- Built-in Data Option-*-*-
-1 - Data For Regression
-2 - Data For Classification
-3 - Data For Clustering
-4 - Data For Dimensional Reduction
-(User) ➜ @Number: 4
-```
-
-You have to choose ***Data For Dimensional Reduction*** and press 4 on your own keyboard. The command line interface would show
-
-```bash
-Successfully loading the built-in data set 'Data_Decomposition.xlsx'.
---------------------
-Index - Column Name
-1 - CITATION
-2 - SAMPLE NAME
-3 - Label
-4 - Notes
-5 - LATITUDE
-6 - LONGITUDE
-7 - Unnamed: 6
-8 - SIO2(WT%)
-9 - TIO2(WT%)
-10 - AL2O3(WT%)
-11 - CR2O3(WT%)
-12 - FEOT(WT%)
-13 - CAO(WT%)
-14 - MGO(WT%)
-15 - MNO(WT%)
-16 - NA2O(WT%)
-17 - Unnamed: 16
-18 - SC(PPM)
-19 - TI(PPM)
-20 - V(PPM)
-21 - CR(PPM)
-22 - NI(PPM)
-23 - RB(PPM)
-24 - SR(PPM)
-25 - Y(PPM)
-26 - ZR(PPM)
-27 - NB(PPM)
-28 - BA(PPM)
-29 - LA(PPM)
-30 - CE(PPM)
-31 - PR(PPM)
-32 - ND(PPM)
-33 - SM(PPM)
-34 - EU(PPM)
-35 - GD(PPM)
-36 - TB(PPM)
-37 - DY(PPM)
-38 - HO(PPM)
-39 - ER(PPM)
-40 - TM(PPM)
-41 - YB(PPM)
-42 - LU(PPM)
-43 - HF(PPM)
-44 - TA(PPM)
-45 - PB(PPM)
-46 - TH(PPM)
-47 - U(PPM)
---------------------
-(Press Enter key to move forward.
-```
-
-Here, we just need to press any keyboard to continue.
-
-```bash
-World Map Projection for A Specific Element Option:
-1 - Yes
-2 - No
-(Plot) ➜ @Number::
-```
-
-We can choose map projection if we need a world map projection for a specific element option. Choose yes, we can choose an element to map. Choose no, skip to the next mode. More information of the map projection can be seen in [map projection](https://pyrolite.readthedocs.io/en/main/installation.html). In this tutorial, we skip it and gain output as:
-
-```bash
--*-*- Data Selected -*-*-
---------------------
-Index - Column Name
-1 - CITATION
-2 - SAMPLE NAME
-3 - Label
-4 - Notes
-5 - LATITUDE
-6 - LONGITUDE
-7 - Unnamed: 6
-8 - SIO2(WT%)
-9 - TIO2(WT%)
-10 - AL2O3(WT%)
-11 - CR2O3(WT%)
-12 - FEOT(WT%)
-13 - CAO(WT%)
-14 - MGO(WT%)
-15 - MNO(WT%)
-16 - NA2O(WT%)
-17 - Unnamed: 16
-18 - SC(PPM)
-19 - TI(PPM)
-20 - V(PPM)
-21 - CR(PPM)
-22 - NI(PPM)
-23 - RB(PPM)
-24 - SR(PPM)
-25 - Y(PPM)
-26 - ZR(PPM)
-27 - NB(PPM)
-28 - BA(PPM)
-29 - LA(PPM)
-30 - CE(PPM)
-31 - PR(PPM)
-32 - ND(PPM)
-33 - SM(PPM)
-34 - EU(PPM)
-35 - GD(PPM)
-36 - TB(PPM)
-37 - DY(PPM)
-38 - HO(PPM)
-39 - ER(PPM)
-40 - TM(PPM)
-41 - YB(PPM)
-42 - LU(PPM)
-43 - HF(PPM)
-44 - TA(PPM)
-45 - PB(PPM)
-46 - TH(PPM)
-47 - U(PPM)
---------------------
-Select the data range you want to process.
-Input format:
-Format 1: "[**, **]; **; [**, **]", such as "[1, 3]; 7; [10, 13]" --> you want to deal with the columns 1, 2, 3, 7, 10, 11, 12, 13
-Format 2: "xx", such as "7" --> you want to deal with the columns 7)
-```
-
-Two options are offered. For PCA, the Format 1 method is more useful in multiple dimensional reduction. As a tutorial, we input ***[10, 15]*** as an example.
-
-**Note: [start_col_num, end_col_num]**
-
-The selected feature information would be given
-
-```bash
-The Selected Data Set:
- AL2O3(WT%) CR2O3(WT%) FEOT(WT%) CAO(WT%) MGO(WT%) MNO(WT%)
-0 3.936000 1.440 3.097000 18.546000 18.478000 0.083000
-1 3.040000 0.578 3.200000 20.235000 17.277000 0.150000
-2 7.016561 NaN 3.172049 20.092611 15.261175 0.102185
-3 3.110977 NaN 2.413834 22.083843 17.349203 0.078300
-4 6.971044 NaN 2.995074 20.530008 15.562149 0.096700
-.. ... ... ... ... ... ...
-104 2.740000 0.060 4.520000 23.530000 14.960000 0.060000
-105 5.700000 0.690 2.750000 20.120000 16.470000 0.120000
-106 0.230000 2.910 2.520000 19.700000 18.000000 0.130000
-107 2.580000 0.750 2.300000 22.100000 16.690000 0.050000
-108 6.490000 0.800 2.620000 20.560000 14.600000 0.070000
-
-[109 rows x 6 columns]
-```
-After continuing with any key, basic information of selected data would be shown
-
-```bash
-Basic Statistical Information:
-
-RangeIndex: 109 entries, 0 to 108
-Data columns (total 6 columns):
- # Column Non-Null Count Dtype
---- ------ -------------- -----
- 0 AL2O3(WT%) 109 non-null float64
- 1 CR2O3(WT%) 98 non-null float64
- 2 FEOT(WT%) 109 non-null float64
- 3 CAO(WT%) 109 non-null float64
- 4 MGO(WT%) 109 non-null float64
- 5 MNO(WT%) 109 non-null float64
-dtypes: float64(6)
-memory usage: 5.2 KB
-None
-Some basic statistic information of the designated data set:
- AL2O3(WT%) CR2O3(WT%) FEOT(WT%) CAO(WT%) MGO(WT%) MNO(WT%)
-count 109.000000 98.000000 109.000000 109.000000 109.000000 109.000000
-mean 4.554212 0.956426 2.962310 21.115756 16.178044 0.092087
-std 1.969756 0.553647 1.133967 1.964380 1.432886 0.054002
-min 0.230000 0.000000 1.371100 13.170000 12.170000 0.000000
-25% 3.110977 0.662500 2.350000 20.310000 15.300000 0.063075
-50% 4.720000 0.925000 2.690000 21.223500 15.920000 0.090000
-75% 6.233341 1.243656 3.330000 22.185450 16.816000 0.110000
-max 8.110000 3.869550 8.145000 25.362000 23.528382 0.400000
-Successfully calculate the pair-wise correlation coefficient among the selected columns.
-Save figure 'Correlation Plot' in C:\Users\74086\output\images\statistic.
-Successfully draw the distribution plot of the selected columns.
-Save figure 'Distribution Histogram' in C:\Users\74086\output\images\statistic.
-Successfully draw the distribution plot after log transformation of the selected columns.
-Save figure 'Distribution Histogram After Log Transformation' in C:\Users\74086\output\images\statistic.
-(Press Enter key to move forward.)
-```
-
-## NAN value process
-
-
-Check the NAN values would be helpful for later analysis. In geochemistrypi frame, this option is finished automatically.
-
-```bash
--*-*- Imputation -*-*-
-Check which column has null values:
---------------------
-AL2O3(WT%) False
-CR2O3(WT%) True
-FEOT(WT%) False
-CAO(WT%) False
-MGO(WT%) False
-MNO(WT%) False
-dtype: bool
---------------------
-The ratio of the null values in each column:
---------------------
-CR2O3(WT%) 0.100917
-AL2O3(WT%) 0.000000
-FEOT(WT%) 0.000000
-CAO(WT%) 0.000000
-MGO(WT%) 0.000000
-MNO(WT%) 0.000000
-dtype: float64
---------------------
-```
-
-Several strategies are offered for processing the missing values, including:
-
-```bash
--*-*- Strategy for Missing Values -*-*-
-1 - Mean Value
-2 - Median Value
-3 - Most Frequent Value
-4 - Constant(Specified Value)
-Which strategy do you want to apply?
-```
-
-We choose the mean Value in this example and the input data be processed automatically as:
-
-```bash
--*-*- Hypothesis Testing on Imputation Method -*-*-
-Null Hypothesis: The distributions of the data set before and after imputing remain the same.
-Thoughts: Check which column rejects null hypothesis.
-Statistics Test Method: kruskal Test
-Significance Level: 0.05
-The number of iterations of Monte Carlo simulation: 100
-The size of the sample for each iteration (half of the whole data set): 54
-Average p-value:
-AL2O3(WT%) 1.0
-CR2O3(WT%) 0.9327453056346102
-FEOT(WT%) 1.0
-CAO(WT%) 1.0
-MGO(WT%) 1.0
-MNO(WT%) 1.0
-Note: 'p-value < 0.05' means imputation method doesn't apply to that column.
-The columns which rejects null hypothesis: None
-Successfully draw the respective probability plot (origin vs. impute) of the selected columns
-Save figure 'Probability Plot' in C:\Users\86188\geopi_output\GeoPi - Rock Classification\Xgboost Algorithm - Test 1\artifacts\image\statistic.
-Successfully store 'Probability Plot' in 'Probability Plot.xlsx' in C:\Users\86188\geopi_output\GeoPi - Rock Classification\Xgboost Algorithm - Test 1\artifacts\image\statistic.
-
-RangeIndex: 109 entries, 0 to 108
-Data columns (total 6 columns):
- # Column Non-Null Count Dtype
---- ------ -------------- -----
- 0 AL2O3(WT%) 109 non-null float64
- 1 CR2O3(WT%) 109 non-null float64
- 2 FEOT(WT%) 109 non-null float64
- 3 CAO(WT%) 109 non-null float64
- 4 MGO(WT%) 109 non-null float64
- 5 MNO(WT%) 109 non-null float64
-dtypes: float64(6)
-memory usage: 5.2 KB
-None
-Some basic statistic information of the designated data set:
- AL2O3(WT%) CR2O3(WT%) FEOT(WT%) CAO(WT%) MGO(WT%) MNO(WT%)
-count 109.000000 109.000000 109.000000 109.000000 109.000000 109.000000
-mean 4.554212 0.956426 2.962310 21.115756 16.178044 0.092087
-std 1.969756 0.524695 1.133967 1.964380 1.432886 0.054002
-min 0.230000 0.000000 1.371100 13.170000 12.170000 0.000000
-25% 3.110977 0.680000 2.350000 20.310000 15.300000 0.063075
-50% 4.720000 0.956426 2.690000 21.223500 15.920000 0.090000
-75% 6.233341 1.170000 3.330000 22.185450 16.816000 0.110000
-max 8.110000 3.869550 8.145000 25.362000 23.528382 0.400000
-Successfully store 'Data Selected Imputed' in 'Data Selected Imputed.xlsx' in C:\Users\86188\geopi_output\GeoPi - Rock Classification\Xgboost Algorithm - Test 1\artifacts\data.
-(Press Enter key to move forward.)
-```
-
-## Feature engineering
-
-
-The next step is the feature engineering options.
-```bash
--*-*- Feature Engineering -*-*-
-The Selected Data Set:
---------------------
-Index - Column Name
-1 - AL2O3(WT%)
-2 - CR2O3(WT%)
-3 - FEOT(WT%)
-4 - CAO(WT%)
-5 - MGO(WT%)
-6 - MNO(WT%)
---------------------
-Feature Engineering Option:
-1 - Yes
-2 - No
-```
-
-Feature engineering options are essential for data analysis. We choose Yes and gain
-
-```bash
--*-*- Feature Engineering -*-*-
-The Selected Data Set:
---------------------
-Index - Column Name
-1 - AL2O3(WT%)
-2 - CR2O3(WT%)
-3 - FEOT(WT%)
-4 - CAO(WT%)
-5 - MGO(WT%)
-6 - MNO(WT%)
---------------------
-Feature Engineering Option:
-1 - Yes
-2 - No
-(Data) ➜ @Number: 1
-Selected data set:
-a - AL2O3(WT%)
-b - CR2O3(WT%)
-c - FEOT(WT%)
-d - CAO(WT%)
-e - MGO(WT%)
-f - MNO(WT%)
-Name the constructed feature (column name), like 'NEW-COMPOUND':
-@input: new Feature
-Build up new feature with the combination of basic arithmatic operators, including '+', '-', '*', '/', '()'.
-Input example 1: a * b - c
---> Step 1: Multiply a column with b column;
---> Step 2: Subtract c from the result of Step 1;
-Input example 2: (d + 5 * f) / g
---> Step 1: Multiply 5 with f;
---> Step 2: Plus d column with the result of Step 1;
---> Step 3: Divide the result of Step 1 by g;
-Input example 3: pow(a, b) + c * d
---> Step 1: Raise the base a to the power of the exponent b;
---> Step 2: Multiply the value of c by the value of d;
---> Step 3: Add the result of Step 1 to the result of Step 2;
-Input example 4: log(a)/b - c
---> Step 1: Take the logarithm of the value a;
---> Step 2: Divide the result of Step 1 by the value of b;
---> Step 3: Subtract the value of c from the result of Step 2;
-You can use mean(x) to calculate the average value.
-@input:
-```
-
-Considering actual need for constructing several new geochemical indexes. We can set up some new indexes. Here, we would set up a new index by *AL2O3/CAO* via keyboard options with *a/d*.
-
-```bash
-Do you want to continue to construct a new feature?
-1 - Yes
-2 - No
-(Data) ➜ @Number: 2
-Successfully store 'Data Before Splitting' in 'Data Before Splitting.xlsx' in C:\Users\74086\output\data.
-Exit Feature Engineering Mode.
-```
-
-## PCA
-
-
-Then we can start PCA by selecting Dimensional Reduction and Principal Component Analysis. The kept component number is a hyper-parameter needs to be decided and here we propose the number is 3. Some PCA information is shown on the window.
-
-```bash
--*-*- Hyper-parameters Specification -*-*-
-Decide the component numbers to keep:
-(Model) ➜ @Number: 3
-*-**-* PCA is running ... *-**-*
-Expected Functionality:
-+ Model Persistence
-+ Principal Components
-+ Explained Variance Ratio
-+ Compositional Bi-plot
-+ Compositional Tri-plot
------* Principal Components *-----
-Every column represents one principal component respectively.
-Every row represents how much that row feature contributes to each principal component respectively.
-The tabular data looks like in format: 'rows x columns = 'features x principal components'.
- PC1 PC2 PC3
-AL2O3(WT%) -0.742029 -0.439057 -0.085773
-CR2O3(WT%) -0.007037 0.082531 -0.213232
-FEOT(WT%) -0.173824 0.219858 0.937257
-CAO(WT%) 0.624609 -0.620584 0.200722
-MGO(WT%) 0.165265 0.605489 -0.168090
-MNO(WT%) -0.003397 0.011160 0.012315
- -0.040834 -0.014650 -0.005382
------* Explained Variance Ratio *-----
-[0.46679568 0.38306839 0.09102234]
------* 2 Dimensions Data Selection *-----
-The software is going to draw related 2d graphs.
-Currently, the data dimension is beyond 2 dimensions.
-Please choose 2 dimensions of the data below.
-1 - PC1
-2 - PC2
-3 - PC3
-Choose dimension - 1 data:
-```
-
-By inputting different component numbers, results of PCA are obtained automatically.
-
-
-Figure 1 PCA Example
-
-
-
-## t-distributed Stochastic Neighbor Embedding (T-SNE)
+## T-distributed Stochastic Neighbor Embedding (T-SNE)
@@ -448,7 +15,7 @@ By inputting different component numbers, results of PCA are obtained automatica
- [5. Model Selection](#5-model-selection)
- [6. T-SNE](#6-t-sne)
-### 1. t-distributed Stochastic Neighbor Embedding (T-SNE)
+### 1. T-distributed Stochastic Neighbor Embedding (T-SNE)
**t-distributed Stochastic Neighbor Embedding** is usually known as T-SNE. T-SNE is an unsupervised learning method, in which the training data we feed to the algorithm does not need the desired labels. T-SNE is a machine learning algorithm used for dimensionality reduction and visualization of high-dimensional data.
It represents the similarity between data points in the high-dimensional space using a Gaussian distribution, creating a probability distribution by measuring this similarity. In the low-dimensional space, T-SNE reconstructs this similarity distribution using the t-distribution. T-SNE aims to preserve the local relationships between data points, ensuring that similar points in the high-dimensional space remain similar in the low-dimensional space.
diff --git a/docs/source/index.rst b/docs/source/index.rst
index 3954b500..845b9322 100644
--- a/docs/source/index.rst
+++ b/docs/source/index.rst
@@ -32,7 +32,6 @@ Welcome to Geochemistry π
Contributing
Local Deployment
Docker Deployment
- Algorithm Functionality Addition
Complete Pull Request
Add New Model To Framework
Docs Link