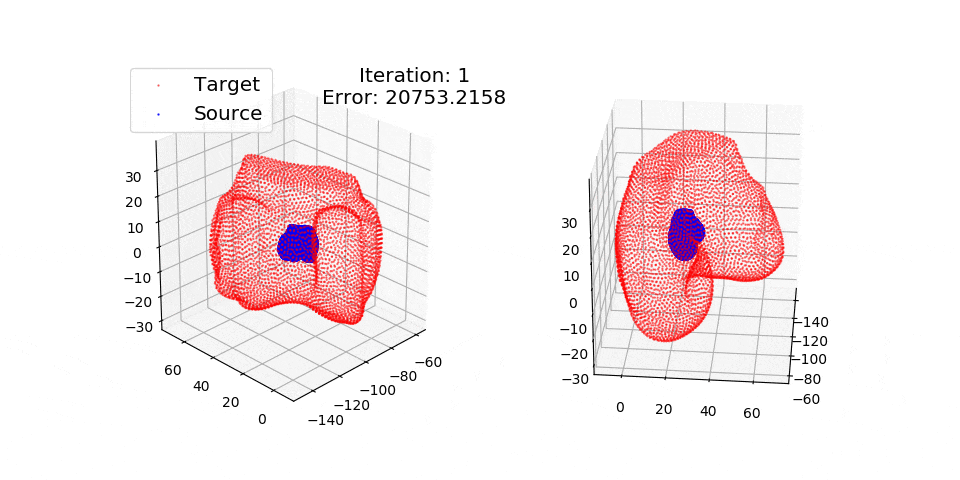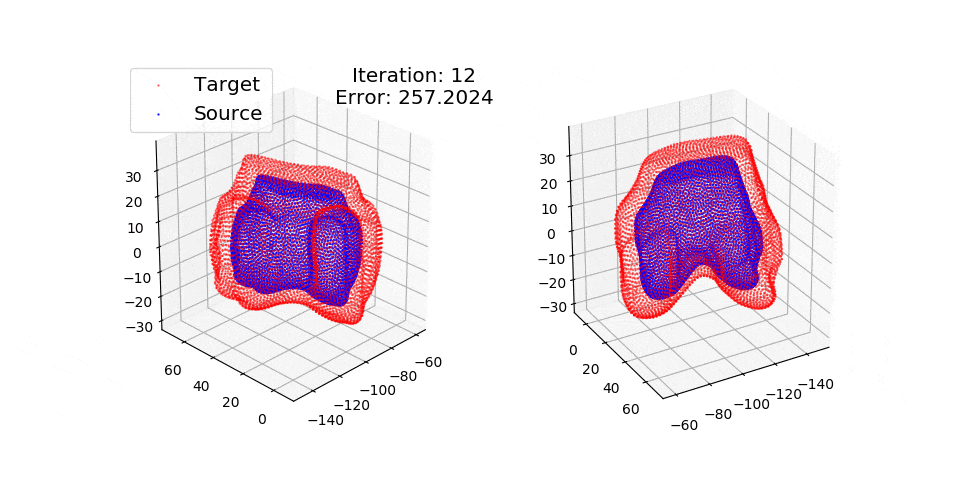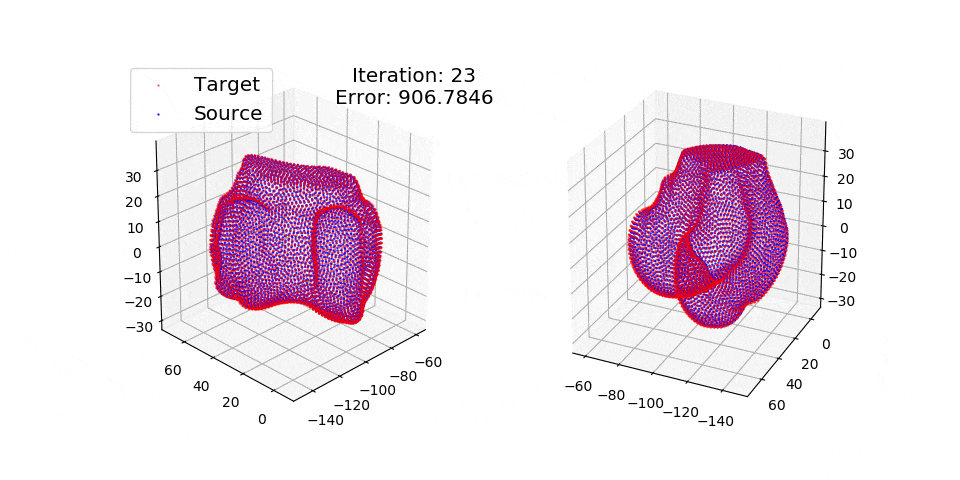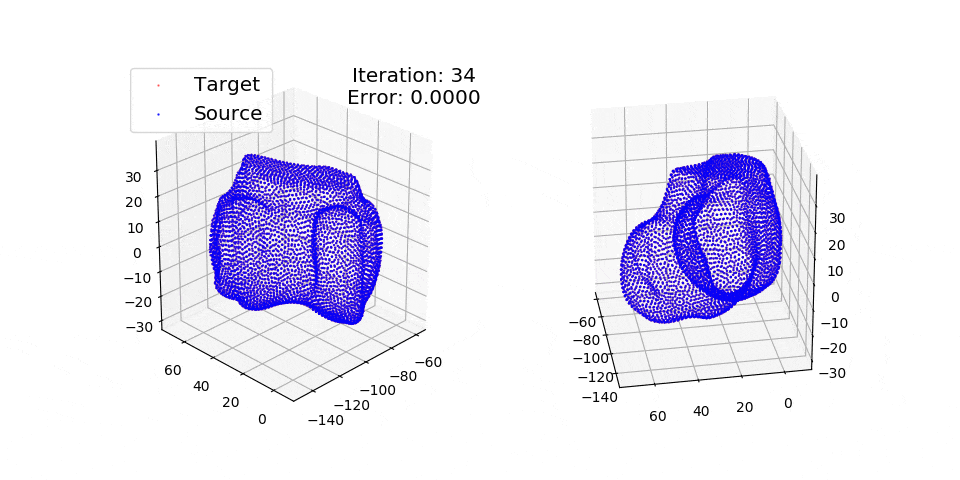
Numpy + Cython Implementation of the Coherent Point Drift Algorithm.
Update to the PyCPD module to include Cython to try and improve performance
Please see here (https://github.com/siavashk/pycpd) for Pure Numpy implementation
Please see here (https://tinyurl.com/tph4u7e) for original manuscript describing CPD
Please see here (https://sites.google.com/site/myronenko/research/cpd) for original code (Matlab) that you can request
Please see here for Matlab code stored on github by secondary source (https://github.com/markeroon/matlab-computer-vision-routines/tree/master/third_party/CoherentPointDrift)
Please see here (https://github.com/gadomski/cpd) for a C++ implementation
This implementation aims to speed up the PyCPD implementation of CPD. First we added cython functions to compute the expectation step of the EM algorithm. This cut E-step times to be ~1/3. E-step is the major bottle neck for rigid and affine registration. Therefore, this function reduces registration of those methods to be ~1/3.
For deformable (non-rigid) registration, the major bottle neck is solving the system of equations for the transformation paramters, which took ~9 seconds (5k point clouds). The first approach we took to speed things up is to implement the low-rank method described in the original CPD paper. This low-rank method significantly reduced computation time and now the entire M-step using default parametrs took <1 second and the E-step is the bottleneck (same as rigid and affine).
The next steps will be to:
- Add the FGT (Fast Gauss Transform). This has the potential to further increase the performance of all methods because it reduces computation for the E-step, which is consistent for all three methods.
- Write more Cython functions to speed up other process/computations.
You should be able to install this by cloning, navigating to this root directory, and installing with pip:
git clone https://github.com/gattia/cycpd
cd cycpd
pip install .
If the above didnt work... previous versions of cycpd had issues with Cython - if you have cython related issues, the following may be helpful:
Must have Cython installed to build package
pip install cythonor
conda install -c anaconda cython
For any operating system you will have to have a C compiler. If you do not have a C compiler you will get errors when building cycpd. You can often follow these errors to install the appropriate packages.
Details about installing C-compiler and other steps necessary for installing Cython can be found here: http://docs.cython.org/en/latest/src/quickstart/install.html. Briefly.
C compiler (gcc) is often present. If it is not, you can install it using:
sudo apt-get install build-essential
You will like need to install gcc (if you havent already). This can be done by installing Apple's xcode command line tools:
xcode-select --install
You will need Visual Studio Community 2019 (free) & Build Tools for Visual Studio 2019. These can be downloaded from: https://visualstudio.microsoft.com/downloads/ You may need newer versions of Visual Studio and it's tools, but thats the one that was required as of writing.
With cython installed:
To run the examples, you will also need matplotlib which is not required for the base package. This can be installed using:
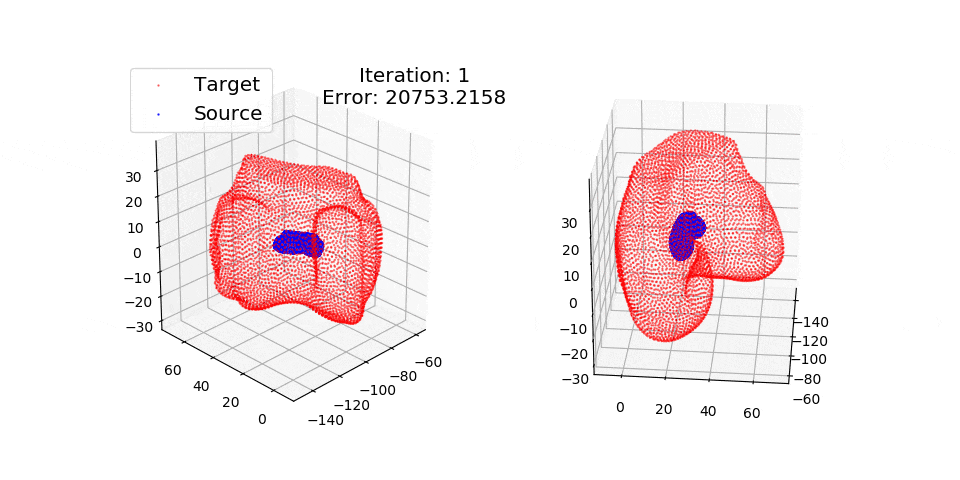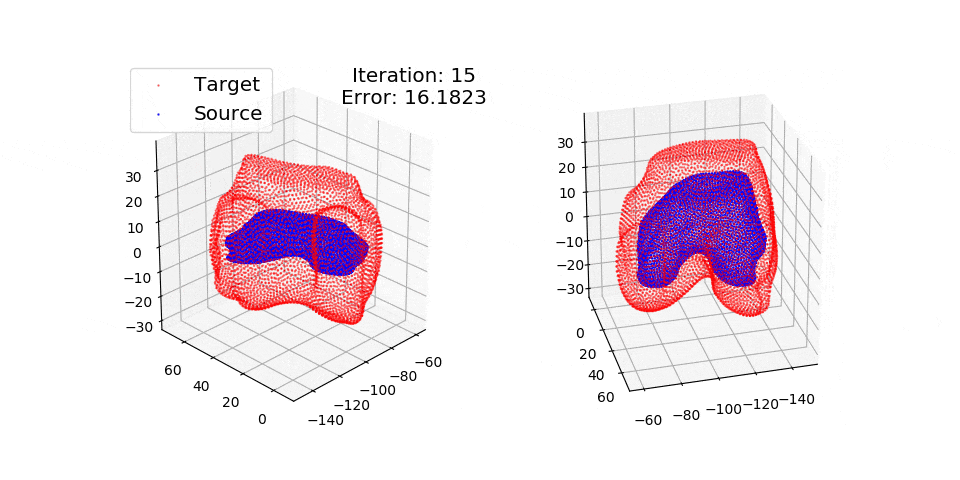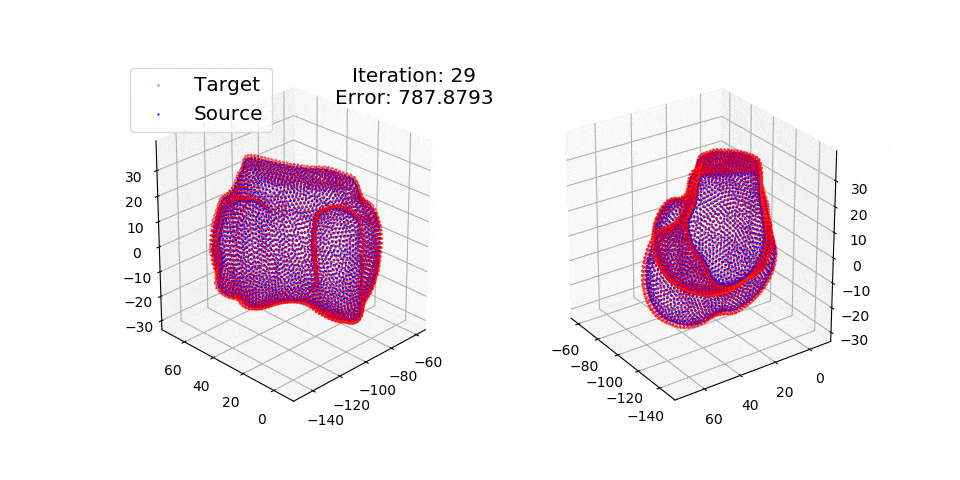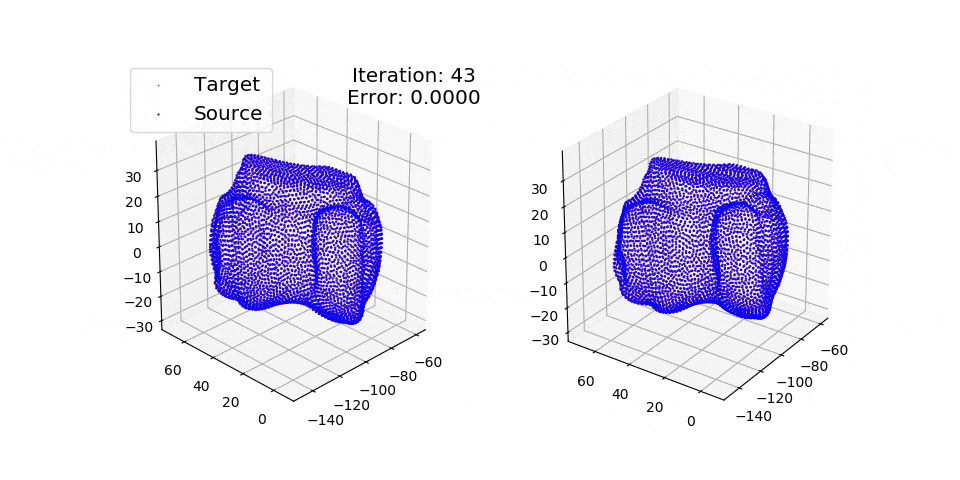
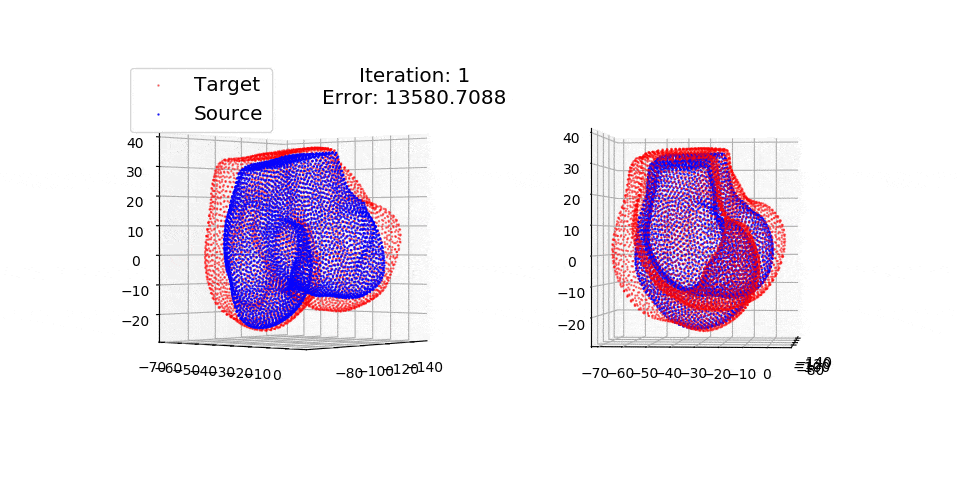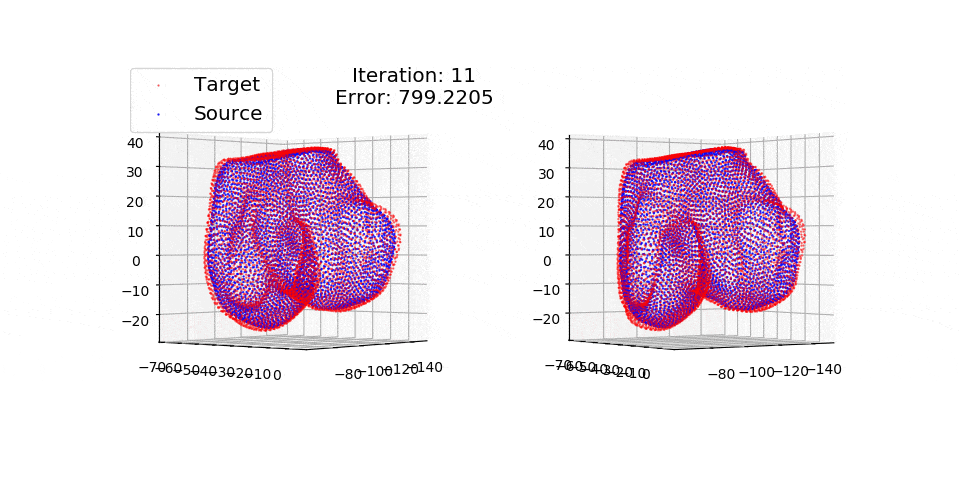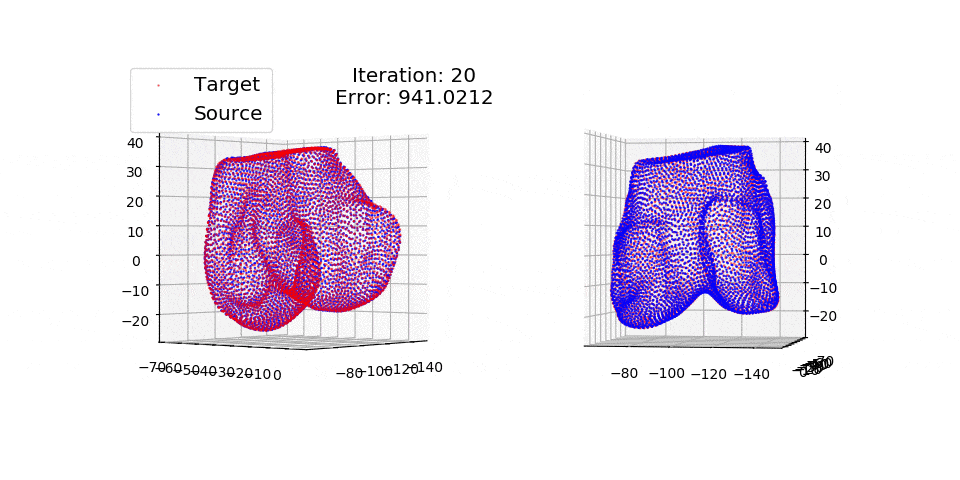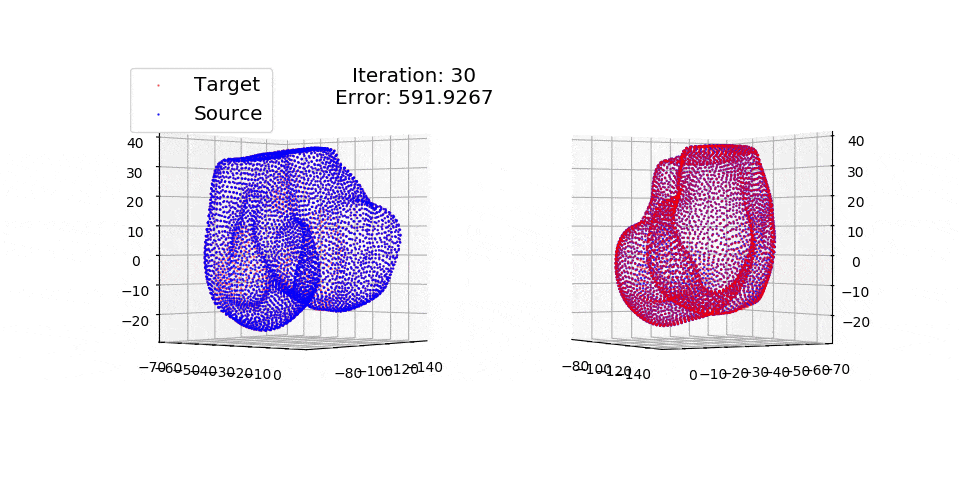
pip install matplotlibThere are three exmples currently implemented. They all show registration of two 3D bones with 5k points each. The Affine applies a transformation to a bone and then use CPD to return it back to its original shape. The rigid and deformable (non-rigid) warps the bone of one person onto a version of that same bone warped to best fit another person. The deformable example will end at 100 epochs (default), at which time it will not have converged fully.
These examples can be run by navigating to the examples folder (after installing) and running:
python knee_rigid_3D.py
python knee_affine_3D.py
python knee_deformable_3D.py
Testing includes rigid, affine, and deformable examples. The rigid, affine, and 2D deformable all test to ensure the algorithm recovers a predefined transformation. The 3D deformable tests to ensure that the resulting registrtaion has errors (between a mesh and the closest point on the other mesh) below a pre-defined tolerance.
These tests are continually run by Github Actions for all new merges/builds. All of these tests can be run by navigating to the cpd directory and running:
pytestInidividual tests can be run by running
python -m pytests path_to_test
path_to_test need be replaced by the path to the approriate test. If in the testing directory, it can be just affine_test.py or similar. If not in testing directory, will need to specify full (absolute or relative) path.
If the test files are run directly, e.g.,:
python affine_test.py
the same tests that are run by pytest will be conducted. Running these files directly will also time the analyses and print the time to do the analysis. Running this way will also run the diagnostics that are built in to the functions and will print those out.
If you want to contribute, please read over the documentaiton in CONTRIBUTING.md
MIT License