library(tidyverse)
-library(stringi)
-library(scales)
-library(gt)library(tidyverse)
+library(stringi)
+library(scales)
+library(gt)library(tidyverse)
-library(stringi)
-library(scales)
-library(gt)library(tidyverse)
+library(stringi)
+library(scales)
+library(gt)I’ll be using the CRAN package repository data returned by tools::CRAN_package_db() to get package and author metadata for the current packages available on CRAN. This returns a data frame with character columns containing most metadata from the DESCRIPTION file of a given R package.
Since this data will change over time, here’s when tools::CRAN_package_db() was run for reference: 2023-05-03.
cran_pkg_db <- tools::CRAN_package_db()
+cran_pkg_db <- tools::CRAN_package_db()
-glimpse(cran_pkg_db)
+glimpse(cran_pkg_db)#> Rows: 19,473
#> Columns: 67
@@ -118,17 +134,57 @@
Wrangle
Since we only care about package and author metadata, a good first step is to remove everything else. This leaves us with a Package field and two author fields: Author and Authors@R. The difference between the two author fields is that Author is an unstructured text field that can contain any text in any format, and Authors@R is a structured text field containing R code that defines authors’ names and roles with the person() function.
-cran_pkg_db <- cran_pkg_db |>
- select(package = Package, authors = Author, authors_r = `Authors@R`) |>
- as_tibble()
+cran_pkg_db <- cran_pkg_db |>
+ select(package = Package, authors = Author, authors_r = `Authors@R`) |>
+ as_tibble()
Here’s a comparison of the two fields, using the dplyr package as an example:
-# Author
-cran_pkg_db |>
- filter(package == "dplyr") |>
- pull(authors) |>
- cat()
+# Author
+cran_pkg_db |>
+ filter(package == "dplyr") |>
+ pull(authors) |>
+ cat()
#> Hadley Wickham [aut, cre] (<https://orcid.org/0000-0003-4757-117X>),
#> Romain François [aut] (<https://orcid.org/0000-0002-2444-4226>),
@@ -137,11 +193,29 @@
#> Davis Vaughan [aut] (<https://orcid.org/0000-0003-4777-038X>),
#> Posit Software, PBC [cph, fnd]
-# Authors@R
-cran_pkg_db |>
- filter(package == "dplyr") |>
- pull(authors_r) |>
- cat()
+# Authors@R
+cran_pkg_db |>
+ filter(package == "dplyr") |>
+ pull(authors_r) |>
+ cat()
#> c(
#> person("Hadley", "Wickham", , "hadley@posit.co", role = c("aut", "cre"),
@@ -179,9 +253,19 @@
From the output above you can see that every package uses the Author field, but not all packages use the Authors@R field. This is unfortunate, because it means that the names and roles of authors need to be extracted from the unstructured text in the Author field for a subset of packages, which is difficult to do and somewhat error-prone. Just for consideration, here’s how many packages don’t use the Authors@R field.
-cran_pkg_db |>
- filter(is.na(authors_r)) |>
- nrow()
+cran_pkg_db |>
+ filter(is.na(authors_r)) |>
+ nrow()
#> [1] 6361
@@ -193,7 +277,43 @@
Extracting from Authors@R
Getting the data we want from the Authors@R field is pretty straightforward. For the packages where this is used, each one has a vector of person objects stored as a character string like:
-mm_string <- "person(\"Michael\", \"McCarthy\", , role = c(\"aut\", \"cre\"))"
+mm_string <- "person(\"Michael\", \"McCarthy\", , role = c(\"aut\", \"cre\"))"
mm_string
@@ -202,18 +322,56 @@
Which can be parsed and evaluated as R code like:
-mm_eval <- eval(parse(text = mm_string))
+mm_eval <- eval(parse(text = mm_string))
-class(mm_eval)
+class(mm_eval)
#> [1] "person"
Then the format() method for the person class can be used to get names and roles into the format I want simply and accurately.
-mm_person <- format(mm_eval, include = c("given", "family"))
-mm_roles <- format(mm_eval, include = c("role"))
-tibble(person = mm_person, roles = mm_roles)
+mm_person <- format(mm_eval, include = c("given", "family"))
+mm_roles <- format(mm_eval, include = c("role"))
+tibble(person = mm_person, roles = mm_roles)
#> # A tibble: 1 × 2
#> person roles
@@ -223,28 +381,130 @@
I’ve wrapped this up into a small helper function, authors_r(), that includes some light tidying steps just to deal with a couple small discrepancies I noticed in a subset of packages.
-# Get names and roles from "person" objects in the Authors@R field
-authors_r <- function(x) {
- # Some light preprocessing is needed to replace the unicode symbol for line
- # breaks with the regular "\n". This is an edge case from at least one
- # package.
- code <- str_replace_all(x, "\\<U\\+000a\\>", "\n")
- persons <- eval(parse(text = code))
- person <- str_trim(format(persons, include = c("given", "family")))
- roles <- format(persons, include = c("role"))
- tibble(person = person, roles = roles)
+# Get names and roles from "person" objects in the Authors@R field
+authors_r <- function(x) {
+ # Some light preprocessing is needed to replace the unicode symbol for line
+ # breaks with the regular "\n". This is an edge case from at least one
+ # package.
+ code <- str_replace_all(x, "\\<U\\+000a\\>", "\n")
+ persons <- eval(parse(text = code))
+ person <- str_trim(format(persons, include = c("given", "family")))
+ roles <- format(persons, include = c("role"))
+ tibble(person = person, roles = roles)
}
Here’s an example of it with dplyr:
-cran_pkg_db |>
- filter(package == "dplyr") |>
- pull(authors_r) |>
- # Normalizing names leads to more consistent results with summary statistics
- # later on, since some people use things like umlauts and accents
- # inconsistently.
- stri_trans_general("latin-ascii") |>
- authors_r()
+cran_pkg_db |>
+ filter(package == "dplyr") |>
+ pull(authors_r) |>
+ # Normalizing names leads to more consistent results with summary statistics
+ # later on, since some people use things like umlauts and accents
+ # inconsistently.
+ stri_trans_general("latin-ascii") |>
+ authors_r()
#> # A tibble: 6 × 2
#> person roles
@@ -274,53 +534,289 @@
Fortunately, regular expressions actually worked pretty well, so this is the solution I settled on. I tried two approaches to this. First I tried to split the names (and roles) up by commas (and eventually other punctuation as I ran into edge cases). This worked alright; there were clearly errors in the data with this method, but since most packages use a simple structure in the Author field it correctly extracted names from most packages.
Second I tried to extract the names (and roles) directly with a regular expression that could match a variety of names. This is the solution I settled on. It still isn’t perfect, but the data is cleaner than with the other method. Regardless, the difference in number of observations between both methods was only in the mid hundreds—so I think any statistics based on this data, although not completely accurate, are still sufficient to get a good idea of the R developer landscape on CRAN.
-# This regex was adapted from <https://stackoverflow.com/a/7654214/16844576>.
-# It's designed to capture a wide range of names, including those with
-# punctuation in them. It's tailored to this data, so I don't know how well
-# it would generalize to other situations, but feel free to try it.
-persons_roles <- r"((\'|\")*[A-Z]([A-Z]+|(\'[A-Z])?[a-z]+|\.)(?:(\s+|\-)[A-Z]([a-z]+|\.?))*(?:(\'?\s+|\-)[a-z][a-z\-]+){0,2}(\s+|\-)[A-Z](\'?[A-Za-z]+(\'[A-Za-z]+)?|\.)(?:(\s+|\-)[A-Za-z]([a-z]+|\.))*(\'|\")*(?:\s*\[(.*?)\])?)"
-# Some packages put the person() code in the wrong field, but it's also
-# formatted incorrectly and throws an error when evaluated, so the best we can
-# do is just extract the whole thing for each person.
-person_objects <- r"(person\((.*?)\))"
+# This regex was adapted from <https://stackoverflow.com/a/7654214/16844576>.
+# It's designed to capture a wide range of names, including those with
+# punctuation in them. It's tailored to this data, so I don't know how well
+# it would generalize to other situations, but feel free to try it.
+persons_roles <- r"((\'|\")*[A-Z]([A-Z]+|(\'[A-Z])?[a-z]+|\.)(?:(\s+|\-)[A-Z]([a-z]+|\.?))*(?:(\'?\s+|\-)[a-z][a-z\-]+){0,2}(\s+|\-)[A-Z](\'?[A-Za-z]+(\'[A-Za-z]+)?|\.)(?:(\s+|\-)[A-Za-z]([a-z]+|\.))*(\'|\")*(?:\s*\[(.*?)\])?)"
+# Some packages put the person() code in the wrong field, but it's also
+# formatted incorrectly and throws an error when evaluated, so the best we can
+# do is just extract the whole thing for each person.
+person_objects <- r"(person\((.*?)\))"
-# Get names and roles from character strings in the Author field
-authors <- function(x) {
- # The Author field is unstructured and there are idiosyncrasies between
- # different packages. The steps here attempt to fix the idiosyncrasies so
- # authors can be extracted with as few errors as possible.
- persons <- x |>
- # Line breaks should be replaced with spaces in case they occur in the
- # middle of a name.
- str_replace_all("\\n|\\<U\\+000a\\>|\\n(?=[:upper:])", " ") |>
- # Periods should always have a space after them so initials will be
- # recognized as part of a name.
- str_replace_all("\\.", "\\. ") |>
- # Commas before roles will keep them from being included in the regex.
- str_remove_all(",(?= \\[)") |>
- # Get persons and their roles.
- str_extract_all(paste0(persons_roles, "|", person_objects)) |>
- unlist() |>
- # Multiple spaces can be replaced with a single space for cleaner names.
- str_replace_all("\\s+", " ")
+# Get names and roles from character strings in the Author field
+authors <- function(x) {
+ # The Author field is unstructured and there are idiosyncrasies between
+ # different packages. The steps here attempt to fix the idiosyncrasies so
+ # authors can be extracted with as few errors as possible.
+ persons <- x |>
+ # Line breaks should be replaced with spaces in case they occur in the
+ # middle of a name.
+ str_replace_all("\\n|\\<U\\+000a\\>|\\n(?=[:upper:])", " ") |>
+ # Periods should always have a space after them so initials will be
+ # recognized as part of a name.
+ str_replace_all("\\.", "\\. ") |>
+ # Commas before roles will keep them from being included in the regex.
+ str_remove_all(",(?= \\[)") |>
+ # Get persons and their roles.
+ str_extract_all(paste0(persons_roles, "|", person_objects)) |>
+ unlist() |>
+ # Multiple spaces can be replaced with a single space for cleaner names.
+ str_replace_all("\\s+", " ")
- tibble(person = persons) |>
- mutate(
- roles = str_extract(person, "\\[(.*?)\\]"),
- person = str_remove(
- str_remove(person, "\\s*\\[(.*?)\\]"),
- "^('|\")|('|\")$" # Some names are wrapped in quotations
+ tibble(person = persons) |>
+ mutate(
+ roles = str_extract(person, "\\[(.*?)\\]"),
+ person = str_remove(
+ str_remove(person, "\\s*\\[(.*?)\\]"),
+ "^('|\")|('|\")$" # Some names are wrapped in quotations
)
)
}
Here’s an example of it with dplyr. If you compare it to the output from authors_r() above you can see the data quality is still good enough for rock ‘n’ roll, but it isn’t perfect; Posit’s roles are no longer defined because the comma in their name cut off the regex before it captured the roles. So there are some edge cases like this that will create measurement error in the person or roles columns, but I don’t think it’s bad enough to invalidate the results.
-cran_pkg_db |>
- filter(package == "dplyr") |>
- pull(authors) |>
- stri_trans_general("latin-ascii") |>
- authors()
+cran_pkg_db |>
+ filter(package == "dplyr") |>
+ pull(authors) |>
+ stri_trans_general("latin-ascii") |>
+ authors()
#> # A tibble: 6 × 2
#> person roles
@@ -340,53 +836,241 @@
Kurt Hornik, Duncan Murdoch and Achim Zeileis published a nice article in The R Journal explaining the roles of R package authors and where they come from. Briefly, they come from the “Relator and Role” codes and terms from MARC (MAchine-Readable Cataloging, Library of Congress, 2012) here: https://www.loc.gov/marc/relators/relaterm.html.
There are a lot of roles there; I just took the ones that were present in the data at the time I wrote this post.
-marc_roles <- c(
- analyst = "anl",
- architecht = "arc",
- artist = "art",
- author = "aut",
- author_in_quotations = "aqt",
- author_of_intro = "aui",
- bibliographic_antecedent = "ant",
- collector = "col",
- compiler = "com",
- conceptor = "ccp",
- conservator = "con",
- consultant = "csl",
- consultant_to_project = "csp",
- contestant_appellant = "cot",
- contractor = "ctr",
- contributor = "ctb",
- copyright_holder = "cph",
- corrector = "crr",
- creator = "cre",
- data_contributor = "dtc",
- degree_supervisor = "dgs",
- editor = "edt",
- funder = "fnd",
- illustrator = "ill",
- inventor = "inv",
- lab_director = "ldr",
- lead = "led",
- metadata_contact = "mdc",
- musician = "mus",
- owner = "own",
- presenter = "pre",
- programmer = "prg",
- project_director = "pdr",
- scientific_advisor = "sad",
- second_party = "spy",
- sponsor = "spn",
- supporting_host = "sht",
- teacher = "tch",
- thesis_advisor = "ths",
- translator = "trl",
- research_team_head = "rth",
- research_team_member = "rtm",
- researcher = "res",
- reviewer = "rev",
- witness = "wit",
- woodcutter = "wdc"
+marc_roles <- c(
+ analyst = "anl",
+ architecht = "arc",
+ artist = "art",
+ author = "aut",
+ author_in_quotations = "aqt",
+ author_of_intro = "aui",
+ bibliographic_antecedent = "ant",
+ collector = "col",
+ compiler = "com",
+ conceptor = "ccp",
+ conservator = "con",
+ consultant = "csl",
+ consultant_to_project = "csp",
+ contestant_appellant = "cot",
+ contractor = "ctr",
+ contributor = "ctb",
+ copyright_holder = "cph",
+ corrector = "crr",
+ creator = "cre",
+ data_contributor = "dtc",
+ degree_supervisor = "dgs",
+ editor = "edt",
+ funder = "fnd",
+ illustrator = "ill",
+ inventor = "inv",
+ lab_director = "ldr",
+ lead = "led",
+ metadata_contact = "mdc",
+ musician = "mus",
+ owner = "own",
+ presenter = "pre",
+ programmer = "prg",
+ project_director = "pdr",
+ scientific_advisor = "sad",
+ second_party = "spy",
+ sponsor = "spn",
+ supporting_host = "sht",
+ teacher = "tch",
+ thesis_advisor = "ths",
+ translator = "trl",
+ research_team_head = "rth",
+ research_team_member = "rtm",
+ researcher = "res",
+ reviewer = "rev",
+ witness = "wit",
+ woodcutter = "wdc"
)
@@ -394,49 +1078,171 @@
Tidying the data
With all the explanations out of the way we can now tidy the data with our helper functions.
-cran_authors <- cran_pkg_db |>
- mutate(
- # Letters with accents, etc. should be normalized so that names including
- # them are picked up by the regex.
- across(c(authors, authors_r), \(.x) stri_trans_general(.x, "latin-ascii")),
- # The extraction functions aren't vectorized so they have to be mapped over.
- # This creates a list column.
- persons = if_else(
- is.na(authors_r),
- map(authors, \(.x) authors(.x)),
- map(authors_r, \(.x) authors_r(.x))
+cran_authors <- cran_pkg_db |>
+ mutate(
+ # Letters with accents, etc. should be normalized so that names including
+ # them are picked up by the regex.
+ across(c(authors, authors_r), \(.x) stri_trans_general(.x, "latin-ascii")),
+ # The extraction functions aren't vectorized so they have to be mapped over.
+ # This creates a list column.
+ persons = if_else(
+ is.na(authors_r),
+ map(authors, \(.x) authors(.x)),
+ map(authors_r, \(.x) authors_r(.x))
)
- ) |>
- select(-c(authors, authors_r)) |>
- unnest(persons) |>
- # If a package only has one author then they must be the author and creator,
- # so it's safe to impute this when it isn't there.
- group_by(package) |>
- mutate(roles = if_else(
- is.na(roles) & n() == 1, "[aut, cre]", roles
- )) |>
- ungroup()
+ ) |>
+ select(-c(authors, authors_r)) |>
+ unnest(persons) |>
+ # If a package only has one author then they must be the author and creator,
+ # so it's safe to impute this when it isn't there.
+ group_by(package) |>
+ mutate(roles = if_else(
+ is.na(roles) & n() == 1, "[aut, cre]", roles
+ )) |>
+ ungroup()
Then add the indicator columns for roles. Note the use of the walrus operator (:=) here to create new columns from the full names of MARC roles on the left side of the walrus, while detecting the MARC codes with str_detect() on the right side. I’m mapping over this because the left side can’t be a vector.
-cran_authors_tidy <- cran_authors |>
- # Add indicator columns for all roles.
- bind_cols(
- map2_dfc(
- names(marc_roles), marc_roles,
- function(.x, .y) {
- cran_authors |>
- mutate(!!.x := str_detect(roles, .y)) |>
- select(!!.x)
+cran_authors_tidy <- cran_authors |>
+ # Add indicator columns for all roles.
+ bind_cols(
+ map2_dfc(
+ names(marc_roles), marc_roles,
+ function(.x, .y) {
+ cran_authors |>
+ mutate(!!.x := str_detect(roles, .y)) |>
+ select(!!.x)
}
)
- ) |>
- # Not everyone's role is known.
- mutate(unknown = is.na(roles))
+ ) |>
+ # Not everyone's role is known.
+ mutate(unknown = is.na(roles))
This all leaves us with a tidy (mostly error free) data frame about R developers and their roles that is ready to explore:
-glimpse(cran_authors_tidy)
+glimpse(cran_authors_tidy)
#> Rows: 52,719
#> Columns: 50
@@ -498,51 +1304,235 @@
R developer statistics
I’ll start with person-level stats, mainly because some of the other stats are further summaries of these statistics. Nothing fancy here, just the number of packages a person has contributed to, role counts, and nominal and percentile rankings. Both the ranking methods used here give every tie the same (smallest) value, so if two people tied for second place both their ranks would be 2, and the next person’s rank would be 4.
-cran_author_pkg_counts <- cran_authors_tidy |>
- group_by(person) |>
- summarise(
- n_packages = n(),
- across(analyst:unknown, function(.x) sum(.x, na.rm = TRUE))
- ) |>
- mutate(
- # Discretizing this for visualization purposes later on
- n_pkgs_fct = case_when(
- n_packages == 1 ~ "One",
- n_packages == 2 ~ "Two",
- n_packages == 3 ~ "Three",
- n_packages >= 4 ~ "Four+"
+cran_author_pkg_counts <- cran_authors_tidy |>
+ group_by(person) |>
+ summarise(
+ n_packages = n(),
+ across(analyst:unknown, function(.x) sum(.x, na.rm = TRUE))
+ ) |>
+ mutate(
+ # Discretizing this for visualization purposes later on
+ n_pkgs_fct = case_when(
+ n_packages == 1 ~ "One",
+ n_packages == 2 ~ "Two",
+ n_packages == 3 ~ "Three",
+ n_packages >= 4 ~ "Four+"
),
- n_pkgs_fct = factor(n_pkgs_fct, levels = c("One", "Two", "Three", "Four+")),
- rank = min_rank(desc(n_packages)),
- percentile = percent_rank(n_packages) * 100,
- .after = n_packages
- ) |>
- arrange(desc(n_packages))
+ n_pkgs_fct = factor(n_pkgs_fct, levels = c("One", "Two", "Three", "Four+")),
+ rank = min_rank(desc(n_packages)),
+ percentile = percent_rank(n_packages) * 100,
+ .after = n_packages
+ ) |>
+ arrange(desc(n_packages))
Here’s an interactive gt table of the person-level stats so you can find yourself, or ask silly questions like how many other authors share a name with you. If you page or search through it you can also get an idea of the data quality (e.g., try “Posit” under the person column and you’ll see that they don’t use a consistent organization name across all packages, which creates some measurement error here).
Code
-cran_author_pkg_counts |>
- select(-n_pkgs_fct) |>
- gt() |>
- tab_header(
- title = "R Developer Contributions",
- subtitle = "CRAN Package Authorships and Roles"
- ) |>
- text_transform(
- \(.x) str_to_title(str_replace_all(.x, "_", " ")),
- locations = cells_column_labels()
- ) |>
- fmt_number(
- columns = percentile
- ) |>
- fmt(
- columns = rank,
- fns = \(.x) label_ordinal()(.x)
- ) |>
- cols_width(everything() ~ px(120)) |>
- opt_interactive(use_sorting = FALSE, use_filters = TRUE)
+cran_author_pkg_counts |>
+ select(-n_pkgs_fct) |>
+ gt() |>
+ tab_header(
+ title = "R Developer Contributions",
+ subtitle = "CRAN Package Authorships and Roles"
+ ) |>
+ text_transform(
+ \(.x) str_to_title(str_replace_all(.x, "_", " ")),
+ locations = cells_column_labels()
+ ) |>
+ fmt_number(
+ columns = percentile
+ ) |>
+ fmt(
+ columns = rank,
+ fns = \(.x) label_ordinal()(.x)
+ ) |>
+ cols_width(everything() ~ px(120)) |>
+ opt_interactive(use_sorting = FALSE, use_filters = TRUE)
@@ -1006,22 +1996,98 @@
Package contributions
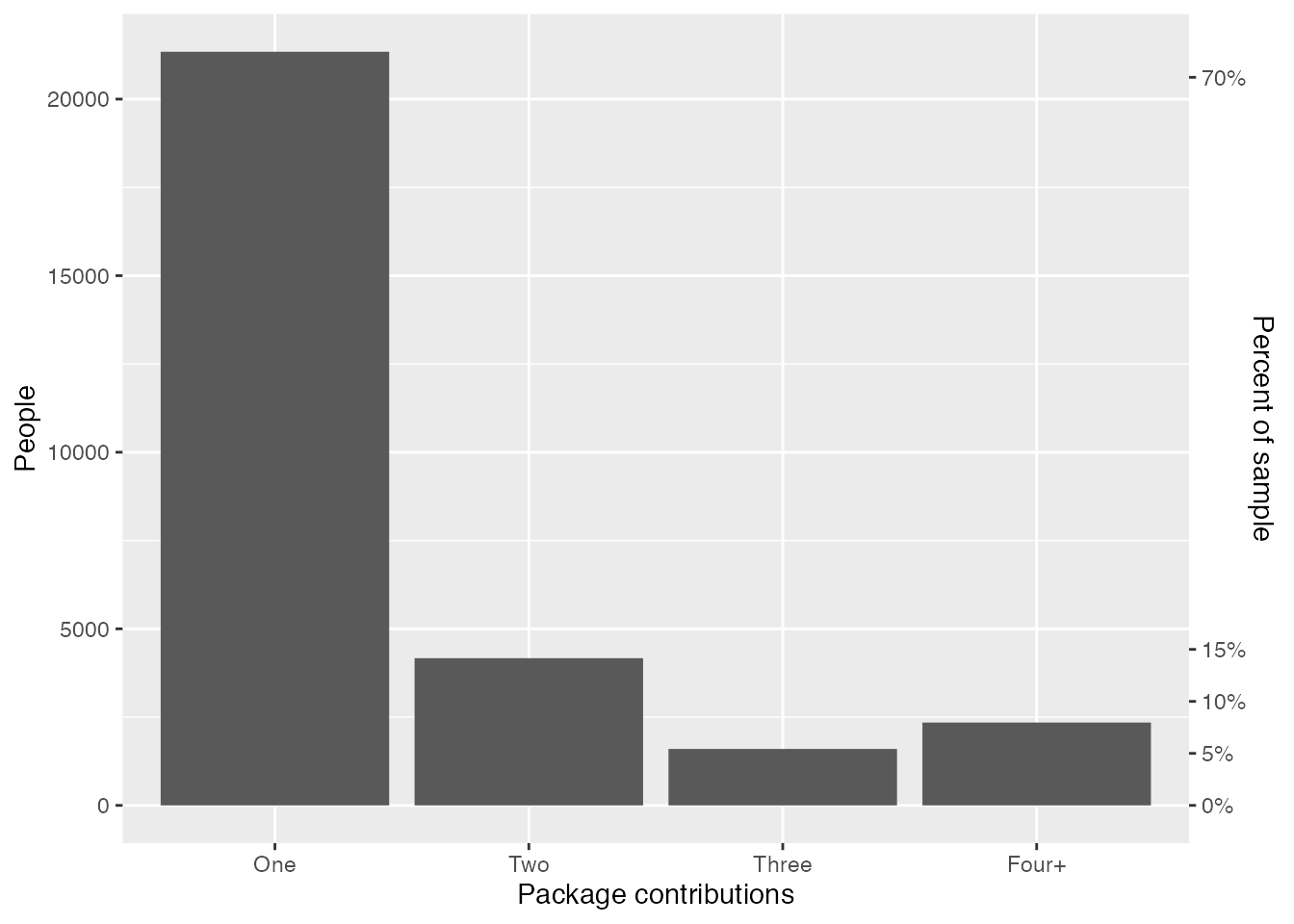
The title of this post probably gave this away, but around 90% of R developers have worked on one to three packages, and only around 10% have worked on four or more packages.
-cran_author_pkg_counts |>
- group_by(n_pkgs_fct) |>
- summarise(n_people = n()) |>
- ggplot(mapping = aes(x = n_pkgs_fct, y = n_people)) +
- geom_col() +
- scale_y_continuous(
- sec.axis = sec_axis(
- trans = \(.x) .x / nrow(cran_author_pkg_counts),
- name = "Percent of sample",
- labels = label_percent(),
- breaks = c(0, .05, .10, .15, .70)
+cran_author_pkg_counts |>
+ group_by(n_pkgs_fct) |>
+ summarise(n_people = n()) |>
+ ggplot(mapping = aes(x = n_pkgs_fct, y = n_people)) +
+ geom_col() +
+ scale_y_continuous(
+ sec.axis = sec_axis(
+ trans = \(.x) .x / nrow(cran_author_pkg_counts),
+ name = "Percent of sample",
+ labels = label_percent(),
+ breaks = c(0, .05, .10, .15, .70)
)
- ) +
- labs(
- x = "Package contributions",
- y = "People"
+ ) +
+ labs(
+ x = "Package contributions",
+ y = "People"
)
@@ -1029,23 +2095,105 @@
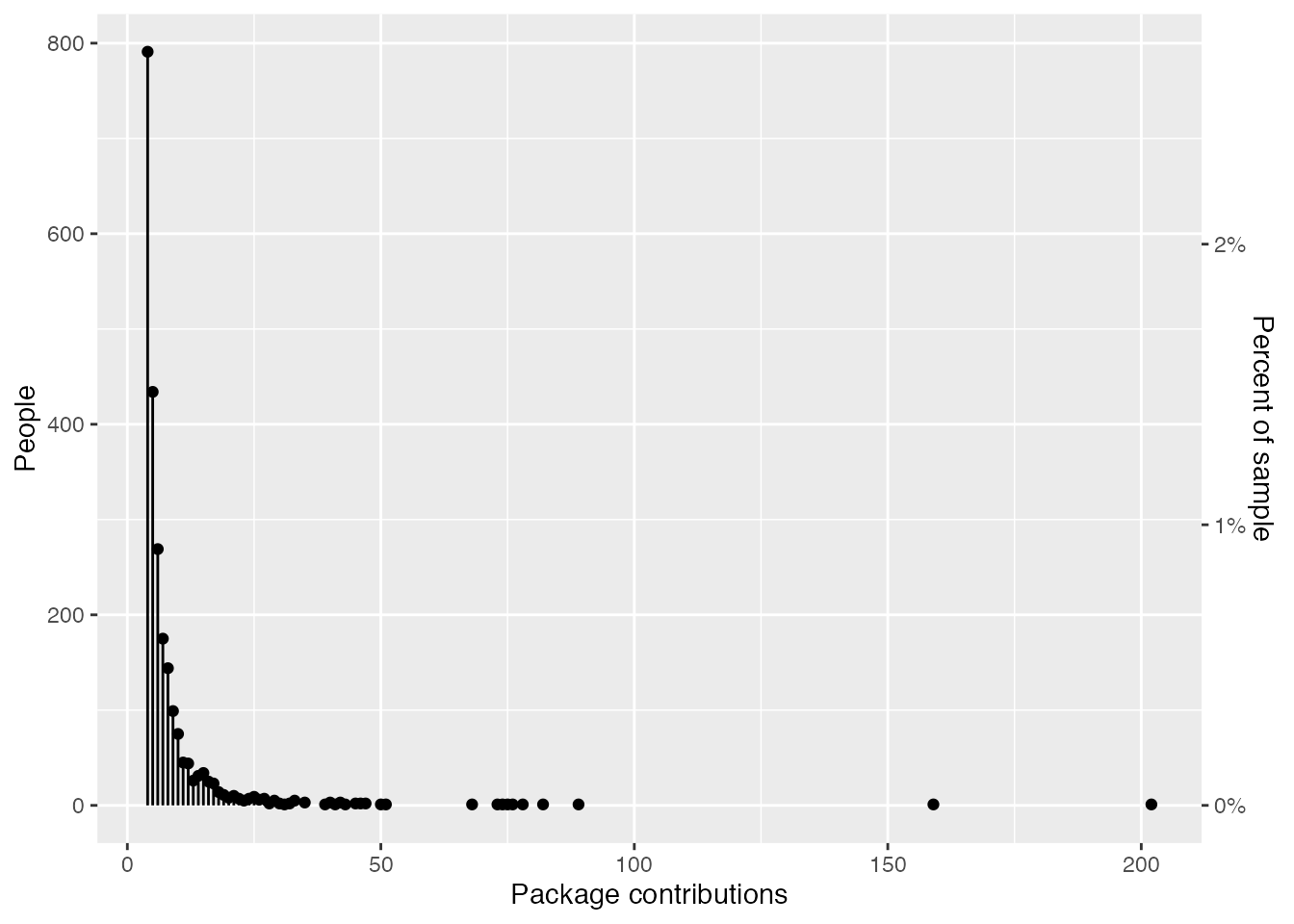
Notably, in the group that have worked on four or more packages, the spread of package contributions is huge. This vast range is mostly driven by people who do R package development as part of their job (e.g., if you look at the cran_author_pkg_counts table above, most of the people at the very top are either professors of statistics or current or former developers from Posit, rOpenSci, or the R Core Team).
-cran_author_pkg_counts |>
- filter(n_pkgs_fct == "Four+") |>
- group_by(rank, n_packages) |>
- summarise(n_people = n()) |>
- ggplot(mapping = aes(x = n_packages, y = n_people)) +
- geom_segment(aes(xend = n_packages, yend = 0)) +
- geom_point() +
- scale_y_continuous(
- sec.axis = sec_axis(
- trans = \(.x) .x / nrow(cran_author_pkg_counts),
- name = "Percent of sample",
- labels = label_percent()
+cran_author_pkg_counts |>
+ filter(n_pkgs_fct == "Four+") |>
+ group_by(rank, n_packages) |>
+ summarise(n_people = n()) |>
+ ggplot(mapping = aes(x = n_packages, y = n_people)) +
+ geom_segment(aes(xend = n_packages, yend = 0)) +
+ geom_point() +
+ scale_y_continuous(
+ sec.axis = sec_axis(
+ trans = \(.x) .x / nrow(cran_author_pkg_counts),
+ name = "Percent of sample",
+ labels = label_percent()
)
- ) +
- labs(
- x = "Package contributions",
- y = "People"
+ ) +
+ labs(
+ x = "Package contributions",
+ y = "People"
)
@@ -1053,15 +2201,51 @@
Here are some subsample summary statistics to compliment the plots above.
-cran_author_pkg_counts |>
- group_by(n_packages >= 4) |>
- summarise(
- n_developers = n(),
- n_pkgs_mean = mean(n_packages),
- n_pkgs_sd = sd(n_packages),
- n_pkgs_median = median(n_packages),
- n_pkgs_min = min(n_packages),
- n_pkgs_max = max(n_packages)
+cran_author_pkg_counts |>
+ group_by(n_packages >= 4) |>
+ summarise(
+ n_developers = n(),
+ n_pkgs_mean = mean(n_packages),
+ n_pkgs_sd = sd(n_packages),
+ n_pkgs_median = median(n_packages),
+ n_pkgs_min = min(n_packages),
+ n_pkgs_max = max(n_packages)
)
#> # A tibble: 2 × 7
@@ -1078,14 +2262,44 @@
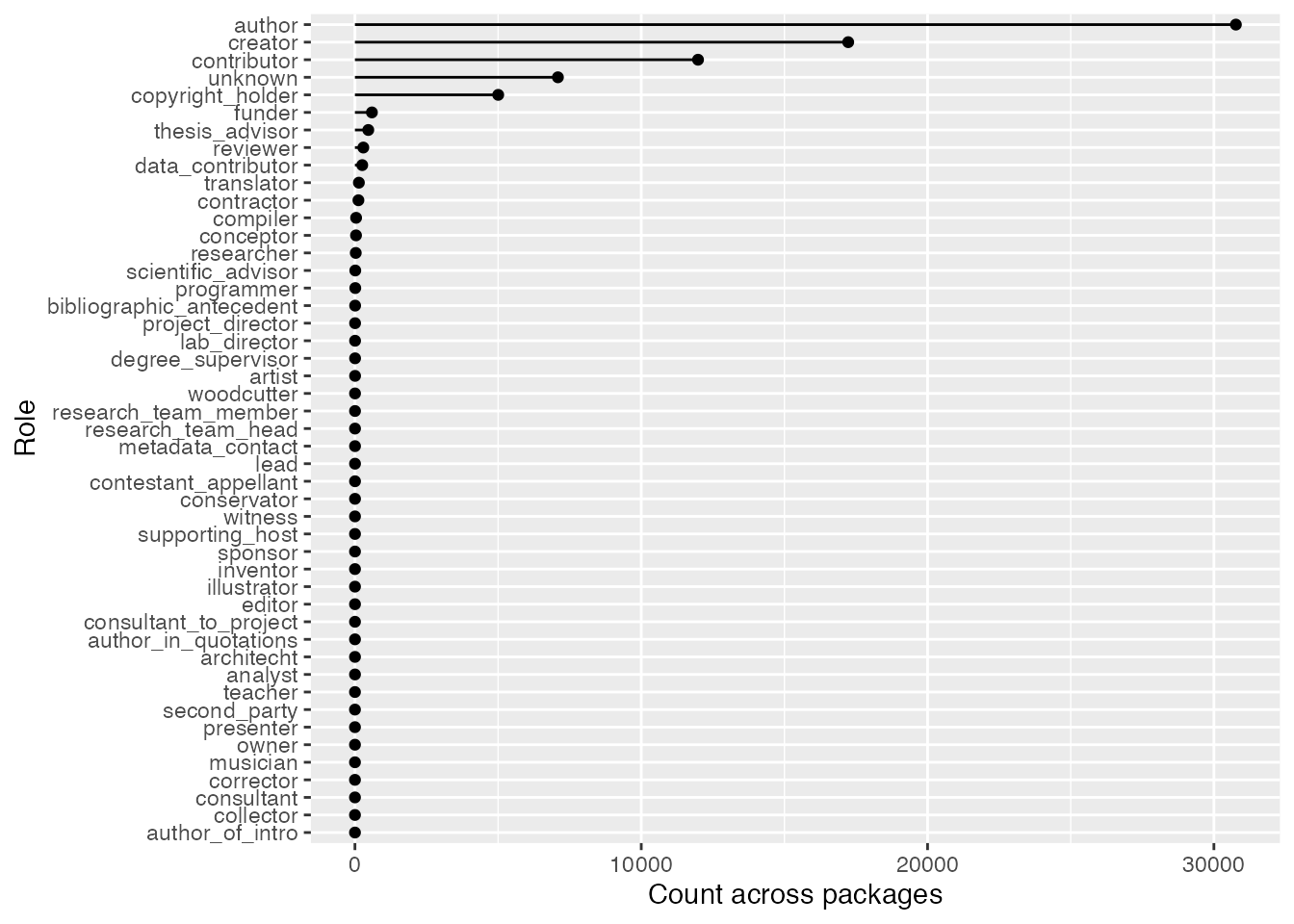
Not every contribution to an R package involves code. For example, two authors of the wiad package were woodcutters! The package is for wood image analysis, so although it’s surprising a role like that exists, it makes a lot of sense in context. Anyways, neat factoids aside, the point of this section is to look at the distribution of different roles in R package development.
To start, let’s get an idea of how many people were involved in programming-related roles. This won’t be universally true, but most of the time the following roles will involve programming:
-programming_roles <-
- c("author", "creator", "contributor", "compiler", "programmer")
+programming_roles <-
+ c("author", "creator", "contributor", "compiler", "programmer")
Here’s the count:
-cran_author_pkg_counts |>
- filter(if_any(!!programming_roles, \(.x) .x > 0)) |>
- nrow()
+cran_author_pkg_counts |>
+ filter(if_any(!!programming_roles, \(.x) .x > 0)) |>
+ nrow()
#> [1] 24170
@@ -1093,16 +2307,96 @@
There were also 5434 whose role was unknown (either because it wasn’t specified or wasn’t picked up by my regex method). Regardless, most people have been involved in programming-related roles, and although other roles occur they’re relatively rare.
Here’s a plot to compliment this point:
-cran_authors_tidy |>
- summarise(across(analyst:unknown, function(.x) sum(.x, na.rm = TRUE))) |>
- pivot_longer(cols = everything(), names_to = "role", values_to = "n") |>
- arrange(desc(n)) |>
- ggplot(mapping = aes(x = n, y = reorder(role, n))) +
- geom_segment(aes(xend = 0, yend = role)) +
- geom_point() +
- labs(
- x = "Count across packages",
- y = "Role"
+cran_authors_tidy |>
+ summarise(across(analyst:unknown, function(.x) sum(.x, na.rm = TRUE))) |>
+ pivot_longer(cols = everything(), names_to = "role", values_to = "n") |>
+ arrange(desc(n)) |>
+ ggplot(mapping = aes(x = n, y = reorder(role, n))) +
+ geom_segment(aes(xend = 0, yend = role)) +
+ geom_point() +
+ labs(
+ x = "Count across packages",
+ y = "Role"
)
@@ -1116,21 +2410,91 @@
This is why Hadley is on the cover of Glamour magazine and we’re not.

-cran_author_pkg_counts |>
- # We don't want organizations or groups here
- filter(!(person %in% c("RStudio", "R Core Team", "Posit Software, PBC"))) |>
- head(20) |>
- select(person, n_packages) |>
- gt() |>
- tab_header(
- title = "Top 20 R Developers",
- subtitle = "Based on number of CRAN package authorships"
- ) |>
- text_transform(
- \(.x) str_to_title(str_replace_all(.x, "_", " ")),
- locations = cells_column_labels()
- ) |>
- cols_width(person ~ px(140))
+cran_author_pkg_counts |>
+ # We don't want organizations or groups here
+ filter(!(person %in% c("RStudio", "R Core Team", "Posit Software, PBC"))) |>
+ head(20) |>
+ select(person, n_packages) |>
+ gt() |>
+ tab_header(
+ title = "Top 20 R Developers",
+ subtitle = "Based on number of CRAN package authorships"
+ ) |>
+ text_transform(
+ \(.x) str_to_title(str_replace_all(.x, "_", " ")),
+ locations = cells_column_labels()
+ ) |>
+ cols_width(person ~ px(140))
@@ -1651,7 +3015,7 @@
-
+
@@ -1666,7 +3030,7 @@
Thanks for reading! I’m Michael, the voice behind Tidy Tales. I am an award winning data scientist and R programmer with the skills and experience to help you solve the problems you care about. You can learn more about me, my consulting services, and my other projects on my personal website.
- Comments
@@ -1675,7 +3039,7 @@
-Session Info
+Session Info
@@ -1733,15 +3097,8 @@ Any of the trademarks, service marks, collective marks, design rights or similar
-
Citation
BibTeX citation:@online{mccarthy2023,
- author = {Michael McCarthy},
- title = {The {Pareto} {Principle} in {R} Package Development},
- date = {2023-05-03},
- url = {https://tidytales.ca/posts/2023-05-03_r-developers},
- langid = {en}
-}
-
For attribution, please cite this work as:
-Michael McCarthy. (2023, May 3). The Pareto Principle in R package
+Citation
For attribution, please cite this work as:
+McCarthy, M. (2023, May 3). The Pareto Principle in R package
development. https://tidytales.ca/posts/2023-05-03_r-developers
.Wrangle
@@ -1766,7 +3123,7 @@ development. h
-
+
-
+
-
+
-
+
-
+
-
+
-
+
-
+
-
+
-
+
-
+
@@ -2012,22 +3369,14 @@ Jan 24, 2023
No matching items
-Artwork
+Artwork
Artwork by Allison Horst.
-Reuse
Citation
BibTeX citation:@online{mccarthy2023,
- author = {Michael McCarthy},
- title = {Reproducible {Data} {Science}},
- date = {2023-01-24},
- url = {https://tidytales.ca/series/2023-01-24_reproducible-data-science},
- langid = {en}
-}
-
For attribution, please cite this work as:
-Michael McCarthy. (2023, January 24). Reproducible Data
-Science. https://tidytales.ca/series/2023-01-24_reproducible-data-science
+Reuse
Citation
For attribution, please cite this work as:
+McCarthy, M. (2023, January 24). Reproducible Data Science. https://tidytales.ca/series/2023-01-24_reproducible-data-science
.Misc
https://tidytales.ca/series/2023-01-24_reproducible-data-science/index.html
@@ -2047,10 +3396,18 @@ Science. Prerequisites
To access the datasets, help pages, and functions that we will use in this code snippet, load the following packages:
-library(ggplot2)
-library(ggdist)
-library(palettes)
-library(forcats)
+library(ggplot2)
+library(ggdist)
+library(palettes)
+library(forcats)
@@ -2109,48 +3466,226 @@ Science.
-
-
-
-
-
Code
-ggplot(likert_scores, aes(x = score, y = item)) +
- stat_slab(
- aes(fill = cut(after_stat(x), breaks = breaks(x))),
- justification = -.2,
- height = 0.7,
- slab_colour = "black",
- slab_linewidth = 0.5,
- trim = TRUE
- ) +
- geom_boxplot(
- width = .2,
- outlier.shape = NA
- ) +
- geom_jitter(width = .1, height = .1, alpha = .3) +
- scale_fill_manual(
- values = pal_ramp(met_palettes$Hiroshige, 5, -1),
- labels = 1:5,
- guide = guide_legend(title = "score", reverse = TRUE)
+ggplot(likert_scores, aes(x = score, y = item)) +
+ stat_slab(
+ aes(fill = cut(after_stat(x), breaks = breaks(x))),
+ justification = -.2,
+ height = 0.7,
+ slab_colour = "black",
+ slab_linewidth = 0.5,
+ trim = TRUE
+ ) +
+ geom_boxplot(
+ width = .2,
+ outlier.shape = NA
+ ) +
+ geom_jitter(width = .1, height = .1, alpha = .3) +
+ scale_fill_manual(
+ values = pal_ramp(met_palettes$Hiroshige, 5, -1),
+ labels = 1:5,
+ guide = guide_legend(title = "score", reverse = TRUE)
)
-ggplot(likert_scores, aes(x = score, y = item)) +
- stat_slab(
- justification = -.2,
- height = 0.7,
- slab_colour = "black",
- slab_linewidth = 0.5,
- trim = FALSE
- ) +
- geom_boxplot(
- width = .2,
- outlier.shape = NA
- ) +
- geom_jitter(width = .1, height = .1, alpha = .3) +
- scale_x_continuous(breaks = 1:5)
+ggplot(likert_scores, aes(x = score, y = item)) +
+ stat_slab(
+ justification = -.2,
+ height = 0.7,
+ slab_colour = "black",
+ slab_linewidth = 0.5,
+ trim = FALSE
+ ) +
+ geom_boxplot(
+ width = .2,
+ outlier.shape = NA
+ ) +
+ geom_jitter(width = .1, height = .1, alpha = .3) +
+ scale_x_continuous(breaks = 1:5)
@@ -2158,7 +3693,7 @@ Science.
-trim = TRUEtrim = TRUE
@@ -2166,7 +3701,7 @@ Science.
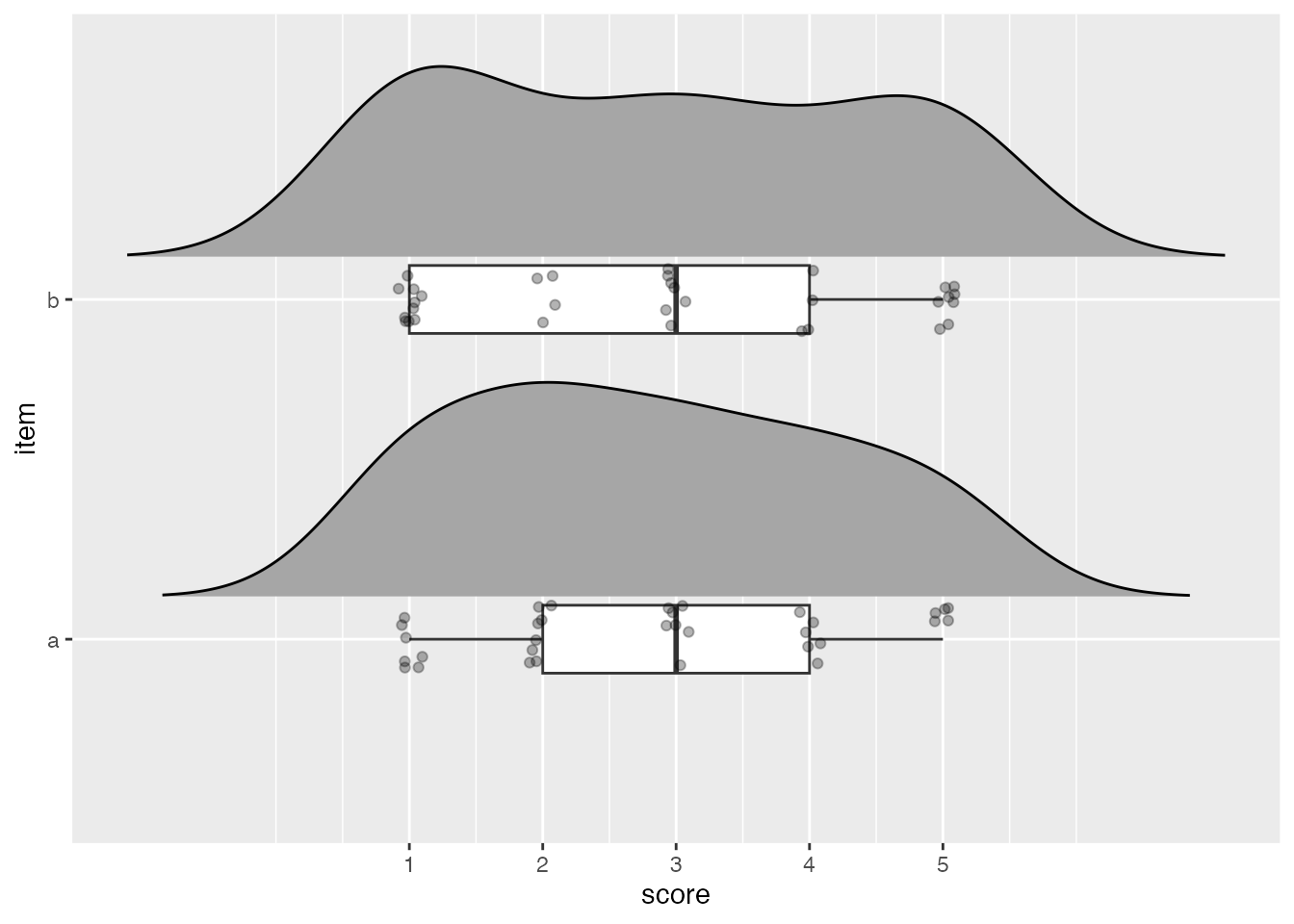
-trim = FALSEtrim = FALSE
First make some data.
-set.seed(123)
+set.seed(123)
-likert_scores <- data.frame(
- item = rep(letters[1:2], times = 33),
- score = sample(1:5, 66, replace = TRUE)
+likert_scores <- data.frame(
+ item = rep(letters[1:2], times = 33),
+ score = sample(1:5, 66, replace = TRUE)
)
It’s straightforward to make density histograms for each item with ggplot2.
-ggplot(likert_scores, aes(x = score, y = after_stat(density))) +
- geom_histogram(
- aes(fill = after_stat(x)),
- bins = 5,
- colour = "black"
- ) +
- scale_fill_gradientn(
- colours = pal_ramp(met_palettes$Hiroshige, 5, -1),
- guide = guide_legend(title = "score", reverse = TRUE)
- ) +
- facet_wrap(vars(fct_rev(item)), ncol = 1)
+ggplot(likert_scores, aes(x = score, y = after_stat(density))) +
+ geom_histogram(
+ aes(fill = after_stat(x)),
+ bins = 5,
+ colour = "black"
+ ) +
+ scale_fill_gradientn(
+ colours = pal_ramp(met_palettes$Hiroshige, 5, -1),
+ guide = guide_legend(title = "score", reverse = TRUE)
+ ) +
+ facet_wrap(vars(fct_rev(item)), ncol = 1)
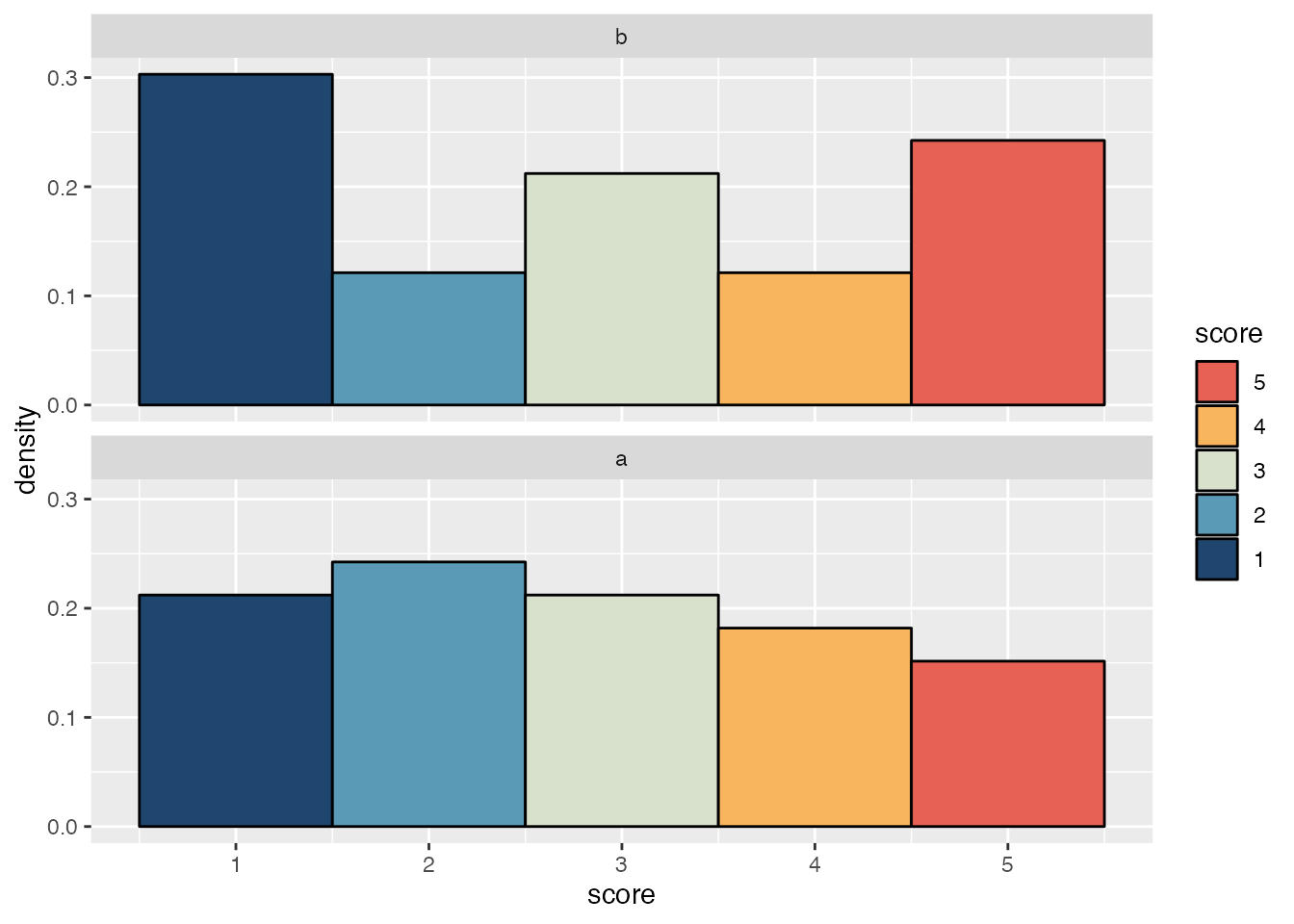
However, the density histograms in this plot can’t be vertically justified to give space for the box and whiskers plot and points used in a typical raincloud plot. For that we need the stat_slab() function from the ggdist package and a small helper function to determine where to put breaks in the histogram.
-#' Set breaks so bins are centred on each score
-#'
-#' @param x A vector of values.
-#' @param width Any value between 0 and 0.5 for setting the width of the bins.
-breaks <- function(x, width = 0.49999999) {
- rep(1:max(x), each = 2) + c(-width, width)
+#' Set breaks so bins are centred on each score
+#'
+#' @param x A vector of values.
+#' @param width Any value between 0 and 0.5 for setting the width of the bins.
+breaks <- function(x, width = 0.49999999) {
+ rep(1:max(x), each = 2) + c(-width, width)
}
The default slab type for stat_slab() is a probability density (or mass) function ("pdf"), but it can also calculate density histograms ("histogram"). To match the appearance of geom_histogram(), the breaks argument needs to be given the location of each bin’s left and right edge; this also necessitates using cut() with the fill aesthetic so the fill breaks correctly align with each bin.
-ggplot(likert_scores, aes(x = score, y = item)) +
- stat_slab(
- # Divide fill into five equal bins
- aes(fill = cut(after_stat(x), breaks = 5)),
- slab_type = "histogram",
- breaks = \(x) breaks(x),
- # Justify the histogram upwards
- justification = -.2,
- # Reduce the histogram's height so it doesn't cover geoms from other items
- height = 0.7,
- # Add black outlines because they look nice
- slab_colour = "black",
- outline_bars = TRUE,
- slab_linewidth = 0.5
- ) +
- geom_boxplot(
- width = .2,
- # Hide outliers since the raw data will be plotted
- outlier.shape = NA
- ) +
- geom_jitter(width = .1, height = .1, alpha = .3) +
- # Cutting the fill into bins puts it on a discrete scale
- scale_fill_manual(
- values = pal_ramp(met_palettes$Hiroshige, 5, -1),
- labels = 1:5,
- guide = guide_legend(title = "score", reverse = TRUE)
+ggplot(likert_scores, aes(x = score, y = item)) +
+ stat_slab(
+ # Divide fill into five equal bins
+ aes(fill = cut(after_stat(x), breaks = 5)),
+ slab_type = "histogram",
+ breaks = \(x) breaks(x),
+ # Justify the histogram upwards
+ justification = -.2,
+ # Reduce the histogram's height so it doesn't cover geoms from other items
+ height = 0.7,
+ # Add black outlines because they look nice
+ slab_colour = "black",
+ outline_bars = TRUE,
+ slab_linewidth = 0.5
+ ) +
+ geom_boxplot(
+ width = .2,
+ # Hide outliers since the raw data will be plotted
+ outlier.shape = NA
+ ) +
+ geom_jitter(width = .1, height = .1, alpha = .3) +
+ # Cutting the fill into bins puts it on a discrete scale
+ scale_fill_manual(
+ values = pal_ramp(met_palettes$Hiroshige, 5, -1),
+ labels = 1:5,
+ guide = guide_legend(title = "score", reverse = TRUE)
)
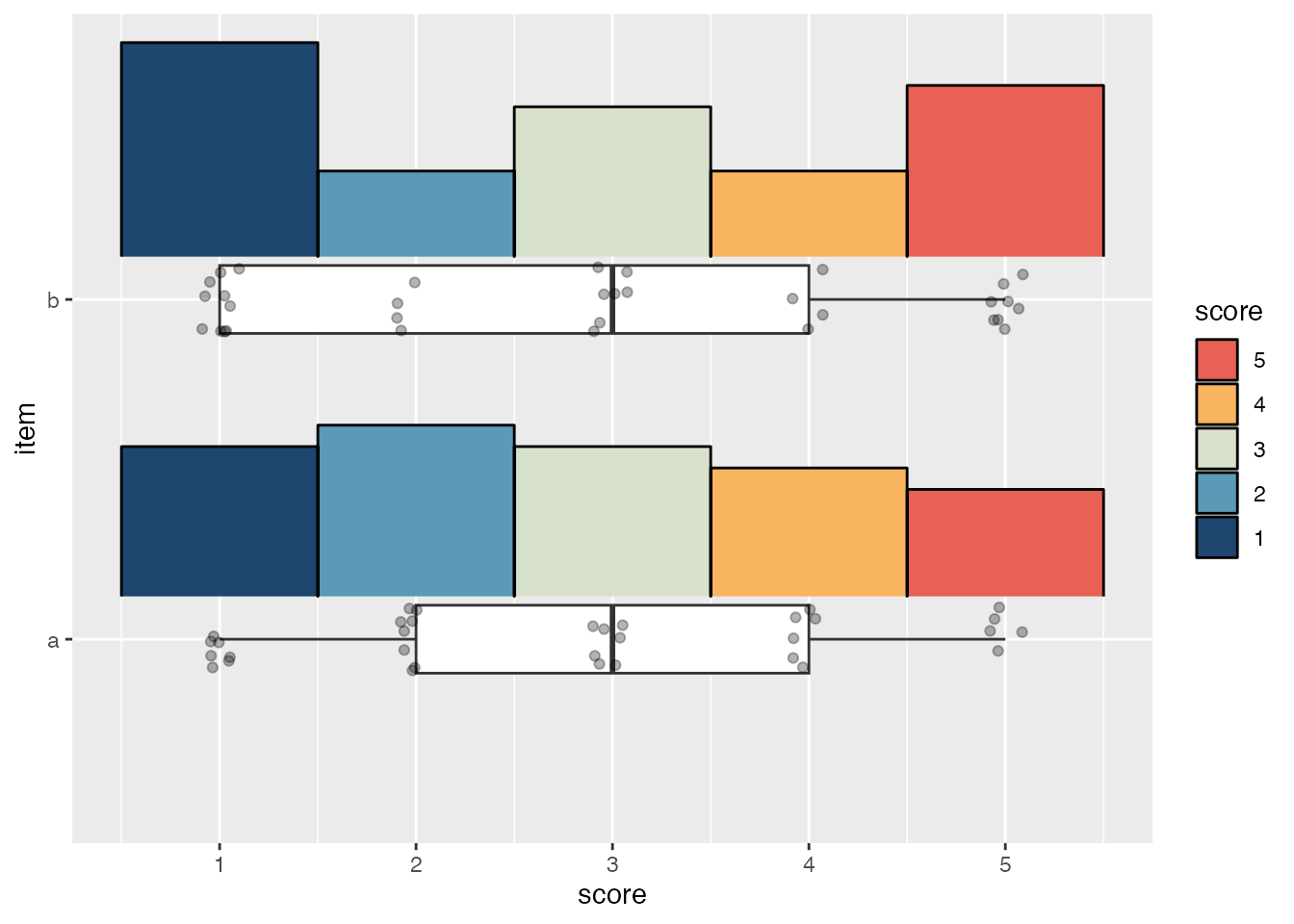
@@ -2253,7 +4058,7 @@ Science.
+
@@ -2268,7 +4073,7 @@ Science. consulting services, and my other projects on my personal website.
- Comments
+Comments
@@ -2277,7 +4082,7 @@ Science. Session Info
+Session Info
@@ -2332,16 +4137,8 @@ Any of the trademarks, service marks, collective marks, design rights or similar
It also makes it difficult to get the fill breaks right, hence the lack of any fill colours in the trim = FALSE example.↩︎
-Reuse
Citation
BibTeX citation:@online{mccarthy2023,
- author = {Michael McCarthy},
- title = {Histogram Raincloud Plots},
- date = {2023-01-19},
- url = {https://tidytales.ca/snippets/2023-01-19_ggdist-histogram-rainclouds},
- langid = {en}
-}
-
For attribution, please cite this work as:
-Michael McCarthy. (2023, January 19). Histogram raincloud
-plots. https://tidytales.ca/snippets/2023-01-19_ggdist-histogram-rainclouds
+Reuse
Citation
For attribution, please cite this work as:
+McCarthy, M. (2023, January 19). Histogram raincloud plots. https://tidytales.ca/snippets/2023-01-19_ggdist-histogram-rainclouds
.Visualize
{ggplot2}
@@ -2362,35 +4159,165 @@ plots. Prerequisites
To access the datasets, help pages, and functions that we will use in this code snippet, load the following packages:
-library(ggplot2)
-library(patchwork)
+library(ggplot2)
+library(patchwork)
Then make some data and ggplot2 plots to be used in the patchwork.
-huron <- data.frame(year = 1875:1972, level = as.vector(LakeHuron))
-h <- ggplot(huron, aes(year))
+huron <- data.frame(year = 1875:1972, level = as.vector(LakeHuron))
+h <- ggplot(huron, aes(year))
-h1 <- h +
- geom_ribbon(aes(ymin = level - 1, ymax = level + 1), fill = "grey70") +
- geom_line(aes(y = level))
+h1 <- h +
+ geom_ribbon(aes(ymin = level - 1, ymax = level + 1), fill = "grey70") +
+ geom_line(aes(y = level))
-h2 <- h + geom_area(aes(y = level))
+h2 <- h + geom_area(aes(y = level))
Comments