-
Notifications
You must be signed in to change notification settings - Fork 31
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Remaining discrepancies A primer on anvi'o
#153
Comments
Inferring taxonomy for metagenomesNote The names in the output of the following two commands is not the same as on the tutorial page command output command output |
Chapter II: Automatic BinningManually curating automatic binning outputsNote Minor note, but I got a slightly different redundancy value than you report in the text ( Text in tutorial
command my output |
|
Thanks for the heads up on these issues @raissameyer :) I started a branch (IGT_fixes_from_Raissa) and fixed the table outputs. I didn't want to update the binning screenshot myself since it has nice colors, so I leave that one to @metehaansever :) |
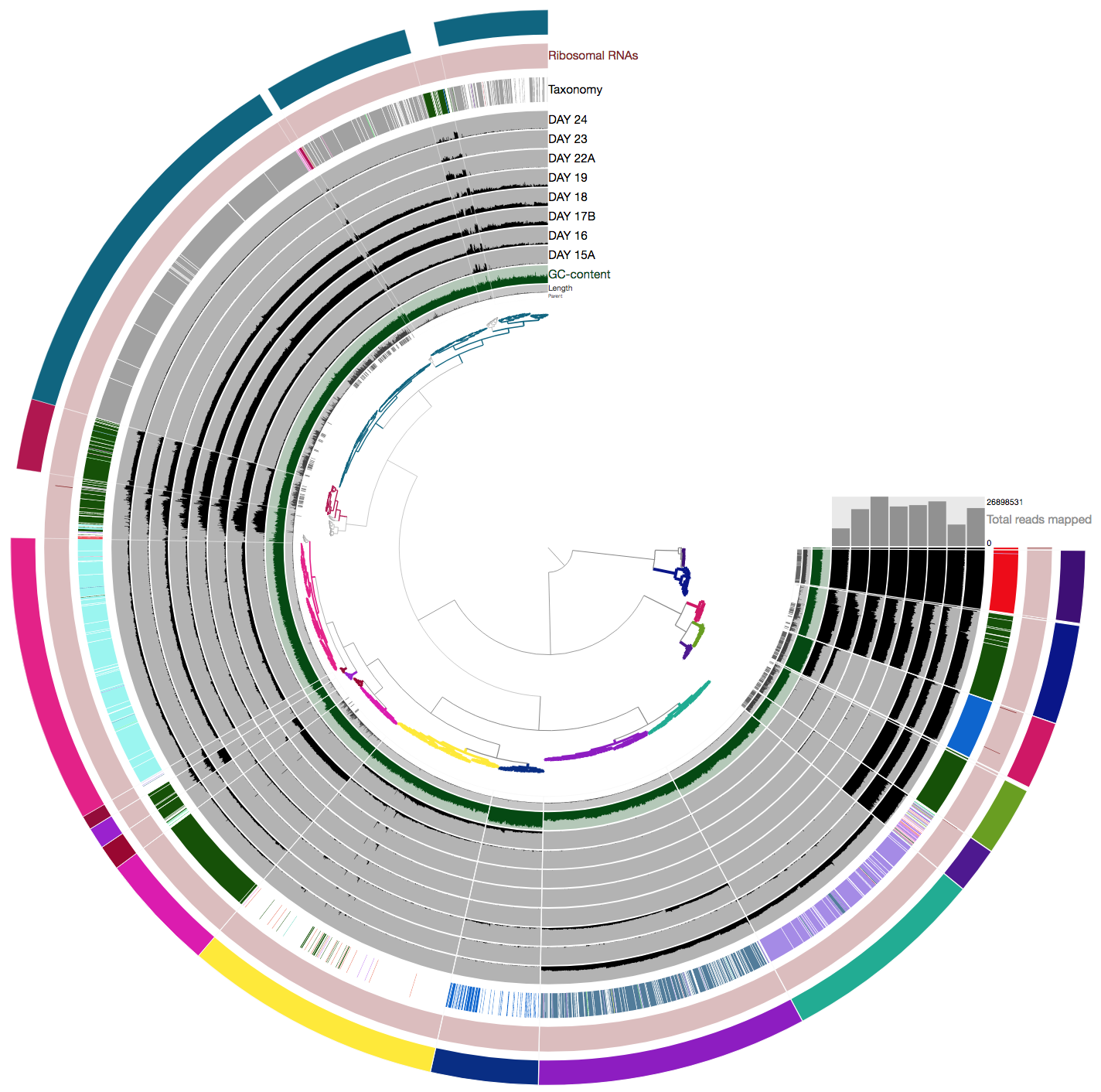
Collection of discrepancies I noticed between the tutorial (after Mete's updates) and my outputs. I'll write one comment per issue as I notice them.
The text was updated successfully, but these errors were encountered: