'
+ for category in categories
+ ]
+ new_index = index_lines[:list_start_index + 1]
+ new_index.extend(categories_entries)
+ new_index.extend(index_lines[list_end_index:])
+ new_index_content = "\n".join(new_index)
+ write_file(path=GALLERY_INDEX_PATH, content=new_index_content)
+
+
+GALLERY_PAGE_TEMPLATE = """
+# Example Workflows - {category}
+
+Below you can find example workflows you can use as inspiration to build your apps.
+
+{examples}
+""".strip()
+
+
+def generate_gallery_page_for_category(
+ category: str,
+ entries: List[WorkflowGalleryEntry],
+) -> None:
+ examples = [
+ generate_gallery_entry_docs(entry=entry)
+ for entry in entries
+ ]
+ page_content = GALLERY_PAGE_TEMPLATE.format(
+ category=category,
+ examples="\n\n".join(examples)
+ )
+ file_path = generate_gallery_page_file_path(category=category)
+ write_file(path=file_path, content=page_content)
+
+
+GALLERY_ENTRY_TEMPLATE = """
+## {title}
+
+{description}
+
+??? tip "Workflow definition"
+
+ ```json
+ {workflow_definition}
+ ```
+""".strip()
+
+
+def generate_gallery_entry_docs(entry: WorkflowGalleryEntry) -> str:
+ return GALLERY_ENTRY_TEMPLATE.format(
+ title=entry.use_case_title,
+ description=entry.use_case_description,
+ workflow_definition="\n\t".join(json.dumps(entry.workflow_definition, indent=4).split("\n")),
+ )
+
+
+def read_file(path: str) -> str:
+ with open(path, "r") as f:
+ return f.read()
+
+
+def write_file(path: str, content: str) -> None:
+ path = os.path.abspath(path)
+ parent_dir = os.path.dirname(path)
+ os.makedirs(parent_dir, exist_ok=True)
+ with open(path, "w") as f:
+ f.write(content)
+
+
+def find_line_with_marker(lines: List[str], marker: str) -> Optional[int]:
+ for i, line in enumerate(lines):
+ if marker in line:
+ return i
+ return None
+
+
+def generate_gallery_page_link(category: str) -> str:
+ file_path = generate_gallery_page_file_path(category=category)
+ return file_path[len(DOCS_ROOT):-3]
+
+
+def generate_gallery_page_file_path(category: str) -> str:
+ category_slug = slugify_category(category=category)
+ return os.path.join(GALLERY_DIR_PATH, f"{category_slug}.md")
+
+
+def slugify_category(category: str) -> str:
+ return category.lower().replace(" ", "_").replace("/", "_")
+
+
+if __name__ == "__main__":
+ generate_gallery()
diff --git a/docs/enterprise/active-learning/active_learning.md b/docs/enterprise/active-learning/active_learning.md
index 512cc72ace..89262d462e 100644
--- a/docs/enterprise/active-learning/active_learning.md
+++ b/docs/enterprise/active-learning/active_learning.md
@@ -36,7 +36,7 @@ Active learning can be disabled by setting `ACTIVE_LEARNING_ENABLED=false` in th
## Usage patterns
Active Learning data collection may be combined with different components of the Roboflow ecosystem. In particular:
-- the `inference` Python package can be used to get predictions from the model and register them at Roboflow platform
+- the `inference` Python package can be used to get predictions from the model and register them on the Roboflow platform
- one may want to use `InferencePipeline` to get predictions from video and register its video frames using Active Learning
- self-hosted `inference` server - where data is collected while processing requests
- Roboflow hosted `inference` - where you let us make sure you get your predictions and data registered. No
diff --git a/docs/foundation/clip.md b/docs/foundation/clip.md
index 632e248716..15499c82e2 100644
--- a/docs/foundation/clip.md
+++ b/docs/foundation/clip.md
@@ -18,7 +18,7 @@ In this guide, we will show:
## How can I use CLIP model in `inference`?
- directly from `inference[clip]` package, integrating the model directly into your code
-- using `inference` HTTP API (hosted locally, or at Roboflow platform), integrating via HTTP protocol
+- using `inference` HTTP API (hosted locally, or on the Roboflow platform), integrating via HTTP protocol
- using `inference-sdk` package (`pip install inference-sdk`) and [`InferenceHTTPClient`](/docs/inference_sdk/http_client.md)
- creating custom code to make HTTP requests (see [API Reference](/api/))
diff --git a/docs/inference_helpers/inference_sdk.md b/docs/inference_helpers/inference_sdk.md
index c1b0c301fc..f94cc0b47d 100644
--- a/docs/inference_helpers/inference_sdk.md
+++ b/docs/inference_helpers/inference_sdk.md
@@ -7,7 +7,7 @@ You can use this client to run models hosted:
1. On the Roboflow platform (use client version `v0`), and;
2. On device with Inference.
-For models trained at Roboflow platform, client accepts the following inputs:
+For models trained on the Roboflow platform, client accepts the following inputs:
- A single image (Given as a local path, URL, `np.ndarray` or `PIL.Image`);
- Multiple images;
@@ -60,7 +60,7 @@ result = loop.run_until_complete(
)
```
-## Configuration options (used for models trained at Roboflow platform)
+## Configuration options (used for models trained on the Roboflow platform)
### configuring with context managers
@@ -195,8 +195,8 @@ Methods that support batching / parallelism:
## Client for core models
-`InferenceHTTPClient` now supports core models hosted via `inference`. Part of the models can be used at Roboflow hosted
-inference platform (use `https://infer.roboflow.com` as url), other are possible to be deployed locally (usually
+`InferenceHTTPClient` now supports core models hosted via `inference`. Part of the models can be used on the Roboflow
+hosted inference platform (use `https://infer.roboflow.com` as url), other are possible to be deployed locally (usually
local server will be available under `http://localhost:9001`).
!!! tip
@@ -705,12 +705,12 @@ to prevent errors)
## Why does the Inference client have two modes (`v0` and `v1`)?
We are constantly improving our `infrence` package - initial version (`v0`) is compatible with
-models deployed at Roboflow platform (task types: `classification`, `object-detection`, `instance-segmentation` and
+models deployed on the Roboflow platform (task types: `classification`, `object-detection`, `instance-segmentation` and
`keypoints-detection`)
are supported. Version `v1` is available in locally hosted Docker images with HTTP API.
Locally hosted `inference` server exposes endpoints for model manipulations, but those endpoints are not available
-at the moment for models deployed at Roboflow platform.
+at the moment for models deployed on the Roboflow platform.
`api_url` parameter passed to `InferenceHTTPClient` will decide on default client mode - URLs with `*.roboflow.com`
will be defaulted to version `v0`.
diff --git a/docs/workflows/about.md b/docs/workflows/about.md
index 0449dfc901..d18d93a867 100644
--- a/docs/workflows/about.md
+++ b/docs/workflows/about.md
@@ -1,23 +1,34 @@
-# Inference Workflows
+# Workflows
-## What is a Workflow?
+## What is Roboflow Workflows?
-Workflows allow you to define multi-step processes that run one or more models to return results based on model outputs and custom logic.
+Roboflow Workflows is an ecosystem that enables users to create machine learning applications using a wide range
+of pluggable and reusable blocks. These blocks are organized in a way that makes it easy for users to design
+and connect different components. Graphical interface allows to visually construct workflows
+without needing extensive technical expertise. Once the workflow is designed, Workflows engine runs the
+application, ensuring all the components work together seamlessly, providing a rapid transition
+from prototype to production-ready solutions, allowing you to quickly iterate and deploy applications.
+
+Roboflow offers a growing selection of workflows blocks, and the community can also create new blocks, ensuring
+that the ecosystem is continuously expanding and evolving. Moreover, Roboflow provides flexible deployment options,
+including on-premises and cloud-based solutions, allowing users to deploy their applications in the environment
+that best suits their needs.
With Workflows, you can:
-- Detect, classify, and segment objects in images.
-- Apply logic filters such as establish detection consensus or filter detections by confidence.
+- Detect, classify, and segment objects in images using state-of-the-art models.
+
- Use Large Multimodal Models (LMMs) to make determinations at any stage in a workflow.
+- Introduce elements of business logic to translate model predictions into your domain language
+
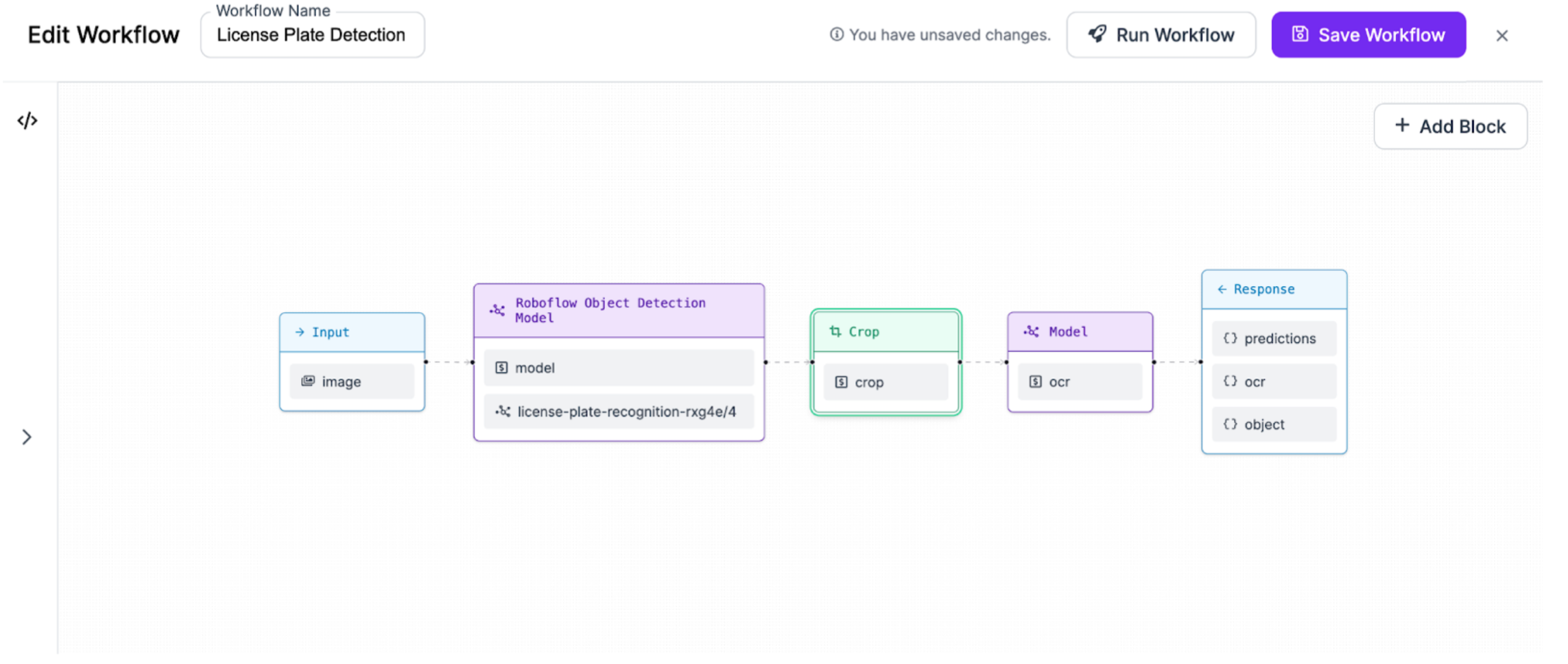
-You can build and configure Workflows in the Roboflow web interface that you can then deploy using the Roboflow Hosted API, self-host locally and on the cloud using inference, or offline to your hardware devices. You can also build more advanced workflows by writing a Workflow configuration directly in the JSON editor.
In this section of documentation, we walk through what you need to know to create and run workflows. Let’s get started!
diff --git a/docs/workflows/blocks_bundling.md b/docs/workflows/blocks_bundling.md
new file mode 100644
index 0000000000..fd565b792b
--- /dev/null
+++ b/docs/workflows/blocks_bundling.md
@@ -0,0 +1,121 @@
+# Bundling Workflows blocks
+
+To efficiently manage the Workflows ecosystem, a standardized method for building and distributing blocks is
+essential. This allows users to create their own blocks and bundle them into Workflow plugins. A Workflow plugin
+is essentially a Python library that implements a defined interface and can be structured in various ways.
+
+This page outlines the mandatory interface requirements and suggests a structure for blocks that aligns with
+the [Workflows versioning](/workflows/versioning) guidelines.
+
+## Proposed structure of plugin
+
+We propose the following structure of plugin:
+
+```
+.
+├── requirements.txt # file with requirements
+├── setup.py # use different package creation method if you like
+├── {plugin_name}
+│ ├── __init__.py # main module that contains loaders
+│ ├── kinds.py # optionally - definitions of custom kinds
+│ ├── {block_name} # package for your block
+│ │ ├── v1.py # version 1 of your block
+│ │ ├── ... # ... next versions
+│ │ └── v5.py # version 5 of your block
+│ └── {block_name} # package for another block
+└── tests # tests for blocks
+```
+
+## Required interface
+
+Plugin must only provide few extensions to `__init__.py` in main package
+compared to standard Python library:
+
+* `load_blocks()` function to provide list of blocks' classes (required)
+
+* `load_kinds()` function to return all custom [kinds](/workflows/kinds/) the plugin defines (optional)
+
+* `REGISTERED_INITIALIZERS` module property which is a dict mapping name of block
+init parameter into default value or parameter-free function constructing that value - optional
+
+
+### `load_blocks()` function
+
+Function is supposed to enlist all blocks in the plugin - it is allowed to define
+a block once.
+
+Example:
+
+```python
+from typing import List, Type
+from inference.core.workflows.prototypes.block import WorkflowBlock
+
+# example assumes that your plugin name is `my_plugin` and
+# you defined the blocks that are imported here
+from my_plugin.block_1.v1 import Block1V1
+from my_plugin.block_2.v1 import Block2V1
+
+def load_blocks() -> List[Type[WorkflowBlock]]:
+ return [
+ Block1V1,
+ Block2V1,
+]
+```
+
+### `load_kinds()` function
+
+`load_kinds()` function to return all custom kinds the plugin defines. It is optional as your blocks
+may not need custom kinds.
+
+Example:
+
+```python
+from typing import List
+from inference.core.workflows.execution_engine.entities.types import Kind
+
+# example assumes that your plugin name is `my_plugin` and
+# you defined the imported kind
+from my_plugin.kinds import MY_KIND
+
+
+def load_kinds() -> List[Kind]:
+ return [MY_KIND]
+```
+
+
+## `REGISTERED_INITIALIZERS` dictionary
+
+As you know from [the docs describing the Workflows Compiler](/workflows/workflows_compiler/)
+and the [blocks development guide](/workflows/create_workflow_block/), Workflow
+blocs are dynamically initialized during compilation and may require constructor
+parameters. Those parameters can default to values registered in the `REGISTERED_INITIALIZERS`
+dictionary. To expose default a value for an init parameter of your block -
+simply register the name of the init param and its value (or a function generating a value) in the dictionary.
+This is optional part of the plugin interface, as not every block requires a constructor.
+
+Example:
+
+```python
+import os
+
+def init_my_param() -> str:
+ # do init here
+ return "some-value"
+
+REGISTERED_INITIALIZERS = {
+ "param_1": 37,
+ "param_2": init_my_param,
+}
+```
+
+## Enabling plugin in your Workflows ecosystem
+
+To load a plugin you must:
+
+* install the Python package with the plugin in the environment you run Workflows
+
+* export an environment variable named `WORKFLOWS_PLUGINS` set to a comma-separated list of names
+of plugins you want to load.
+
+ * Example: to load two plugins `plugin_a` and `plugin_b`, you need to run
+ `export WORKFLOWS_PLUGINS="plugin_a,plugin_b"`
diff --git a/docs/workflows/blocks_connections.md b/docs/workflows/blocks_connections.md
new file mode 100644
index 0000000000..ae53ae73a7
--- /dev/null
+++ b/docs/workflows/blocks_connections.md
@@ -0,0 +1,75 @@
+# Rules dictating which blocks can be connected
+
+A natural question you might ask is: *How do I know which blocks to connect to achieve my desired outcome?*
+This is a crucial question, which is why we've created auto-generated
+[documentation for all supported Workflow blocks](/workflows/blocks/). In this guide, we’ll show you how to use
+these docs effectively and explain key details that help you understand why certain connections between
+blocks are possible, while others may not be.
+
+!!! Note
+
+ Using the Workflows UI in the Roboflow APP you may find compatible connections between steps found
+ automatically without need for your input. This page explains briefly how to deduce if two
+ blocks can be connected, making it possible to connect steps manually if needed. Logically,
+ the page must appear before a link to [blocks gallery](/workflows/blocks/), as it explains
+ how to effectively use these docs. At the same time, it introduces references to concepts
+ further explained in the User and Developer Guide. Please continue reading those sections
+ if you find some concepts presented here needing further explanation.
+
+
+## Navigation the blocks documentation
+
+When you open the blocks documentation, you’ll see a list of all blocks supported by Roboflow. Each block entry
+includes a name, brief description, category, and license for the block. You can click on any block to see more
+detailed information.
+
+On the block details page, you’ll find documentation for all supported versions of that block,
+starting with the latest version. For each version, you’ll see:
+
+- detailed description of the block
+
+- type identifier, which is required in Workflow definitions to identify the specific block used for a step
+
+- table of configuration properties, listing the fields that can be specified in a Workflow definition,
+including their types, descriptions, and whether they can accept a dynamic selector or just a fixed value.
+
+- Available connections, showing which blocks can provide inputs to this block and which can use its outputs.
+
+- A list of input and output bindings:
+
+ - input bindings are the names of step definition properties that can hold selectors, along with the type
+ (or `kind`) of data they pass.
+
+ - output bindings are names and kinds for block outputs that can be used as inputs by steps defined in
+ Workflow definition
+
+- An example of a Workflow step based on the documented block.
+
+The `kind` mentioned above refers to the type of data flowing through the connection during execution,
+and this is further explained in the developer guide.
+
+## What makes connections valid?
+
+Each block provides a manifest that lists the fields to be included in the Workflow Definition when creating a step.
+The Values of these fields in a Workflow Definition may contain:
+
+- References ([selectors](/workflows/definitions/)) to data the block will process, such as step outputs or
+[batch-oriented workflow inputs](/workflows/workflow_execution)
+
+- Configuration values: Specific settings for the step or references ([selectors](/workflows/definitions/)) that
+provide configuration parameters dynamically during execution.
+
+The manifest also includes the block's outputs.
+
+For each step definition field (if it can hold a [selector](/workflows/definitions/)) and step output,
+the expected [kind](/workflows/kinds) is specified. A [kind](/workflows/kinds) is a high-level definition
+of the type of data that will be passed during workflow execution. Simply put, it describes the data that
+will replace the [selector](/workflows/definitions/) during block execution.
+
+To ensure steps are correctly connected, the Workflow Compiler checks if the input and output [kinds](/workflows/kinds)
+match. If they do, the connection is valid.
+
+Additionally, the [`dimensionality level`](/workflows/workflow_execution#dimensionality-level) of the data is considered when
+validating connections. This ensures that data from multiple sources is compatible across the entire Workflow,
+not just between two connected steps. More details on dimensionality levels can be found in the
+[user guide describing workflow execution](/workflows/workflow_execution).
\ No newline at end of file
diff --git a/docs/workflows/create_and_run.md b/docs/workflows/create_and_run.md
index d562475483..1669cad929 100644
--- a/docs/workflows/create_and_run.md
+++ b/docs/workflows/create_and_run.md
@@ -1,220 +1,161 @@
# How to Create and Run a Workflow
-## Example (Web App)
+In this example, we are going to build a Workflow from scratch that detects dogs, classifies their breeds and
+visualizes results.
-In this example, we are going to build a Workflow from scratch that detects license plates, crops the license plate, and then runs OCR on the license plate.
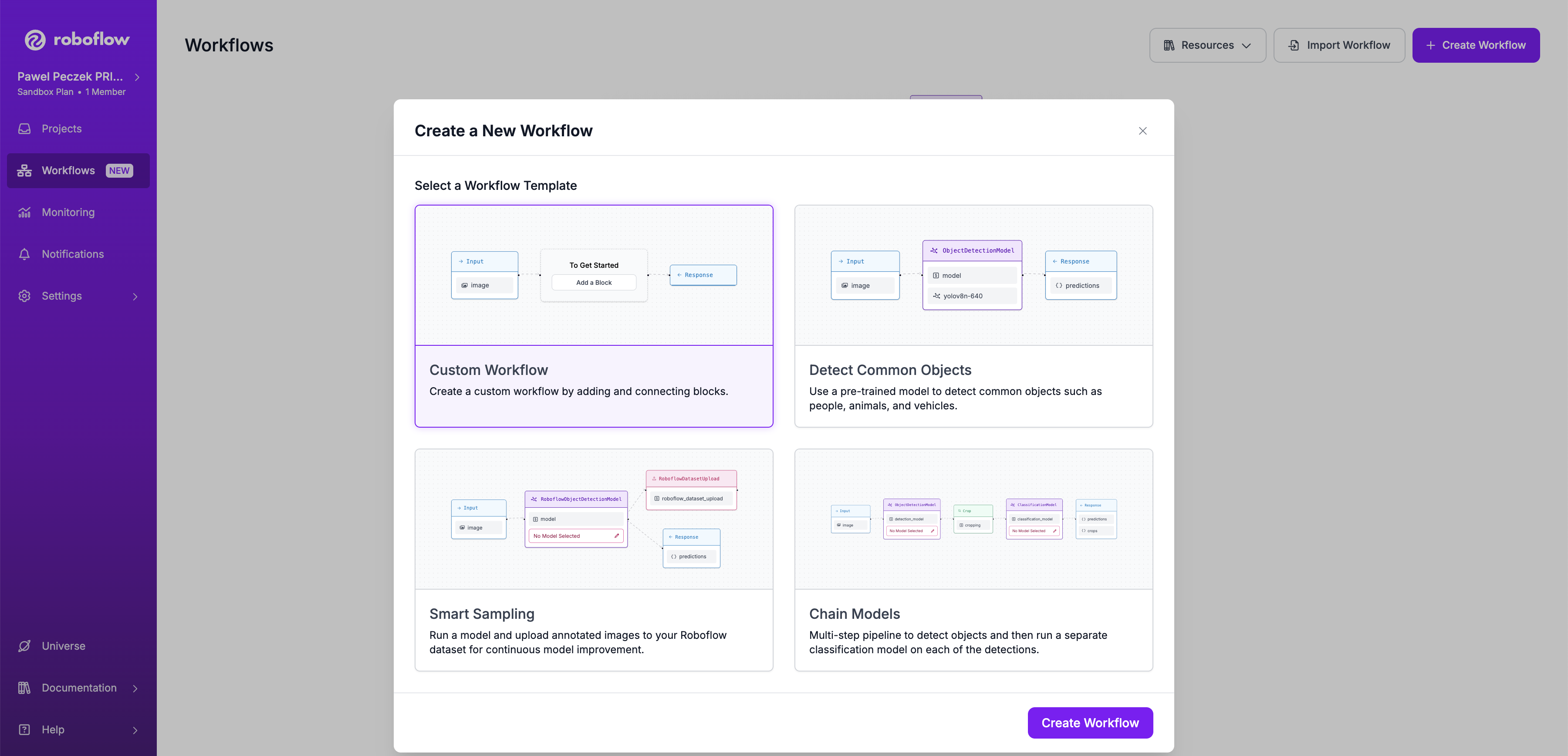
+## Step 1: Create a Workflow
-### Step 1: Create a Workflow
+Open [https://app.roboflow.com/](https://app.roboflow.com/) in your browser, and navigate to the Workflows tab to click
+the Create Workflows button. Select Custom Workflow to start the creation process.
-Navigate to the Workflows tab at the top of your workspace and select the Create Workflows button. We are going to start with a Single Model Workflow.
+
-
-### Step 2: Add Crop
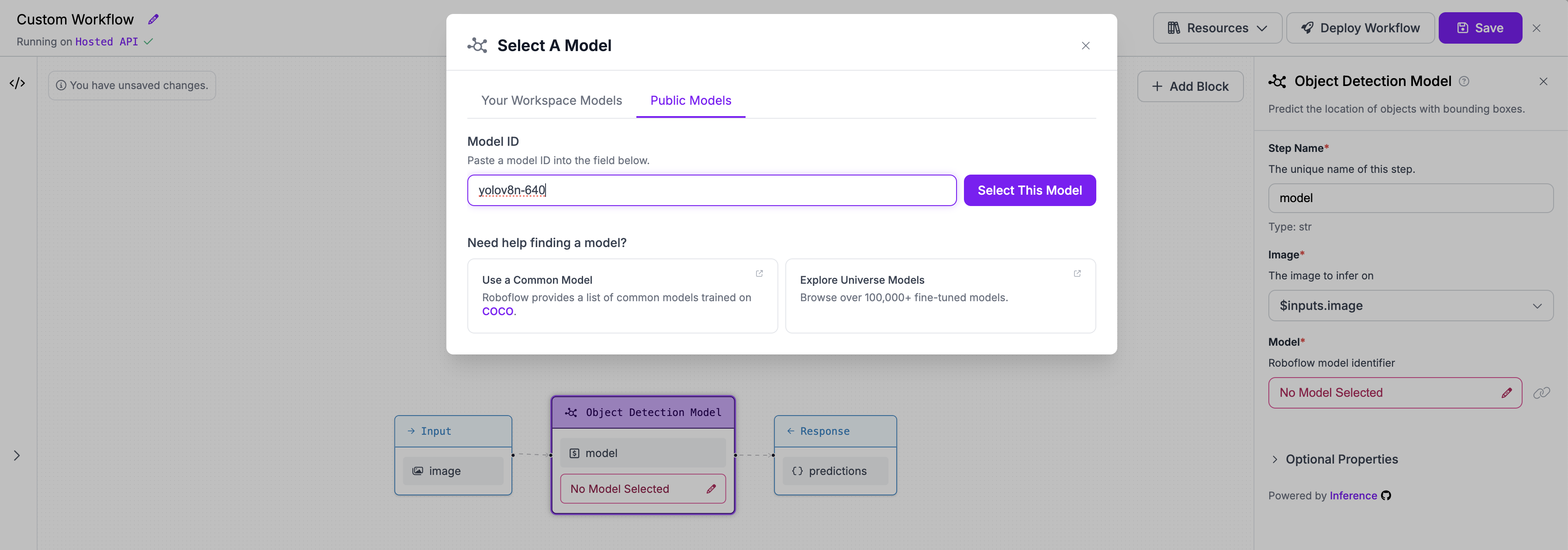
+## Step 2: Add an object detection model
+We need to add a block with an object detection model to the existing workflow. We will use the `yolov8n-640` model.
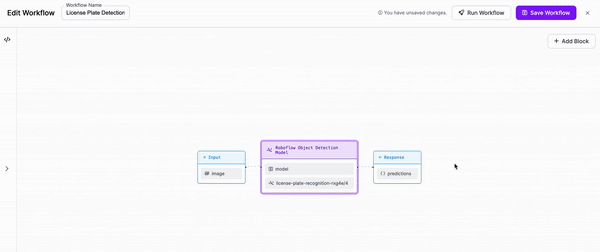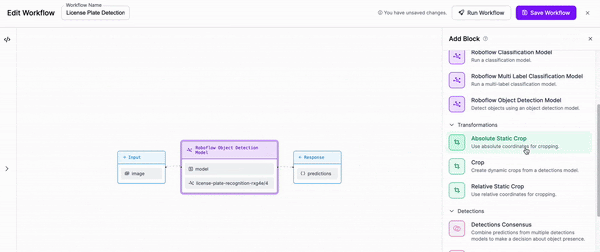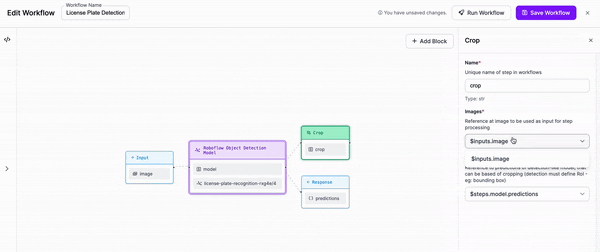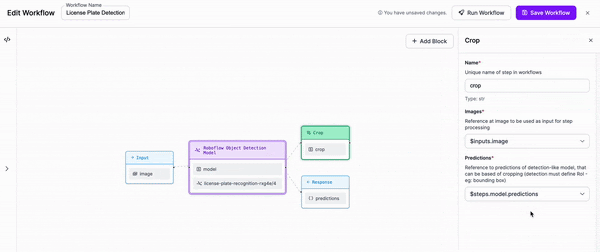
-Next, we are going to add a block to our Workflow that crops the objects that our first model detects.
-
-
-
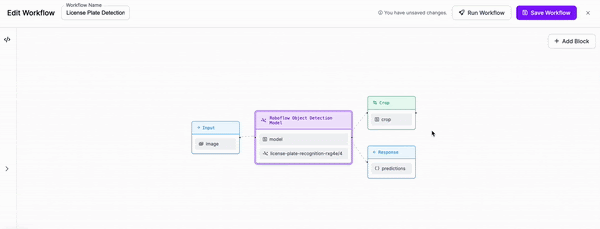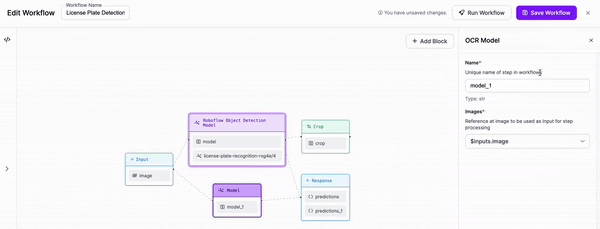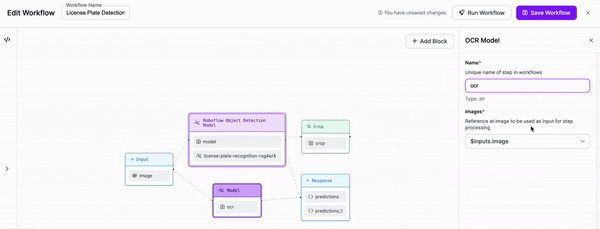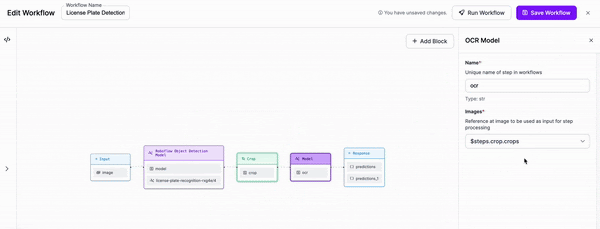
-### Step 3: Add OCR
+
-We are then going to add an OCR model for text recognition to our Workflow. We will need to adjust the parameters in order to set the cropped object from our previous block as the input for this block.
+## Step 3: Crop each detected object to run breed classification
-
-
-### Step 4: Add outputs to our response
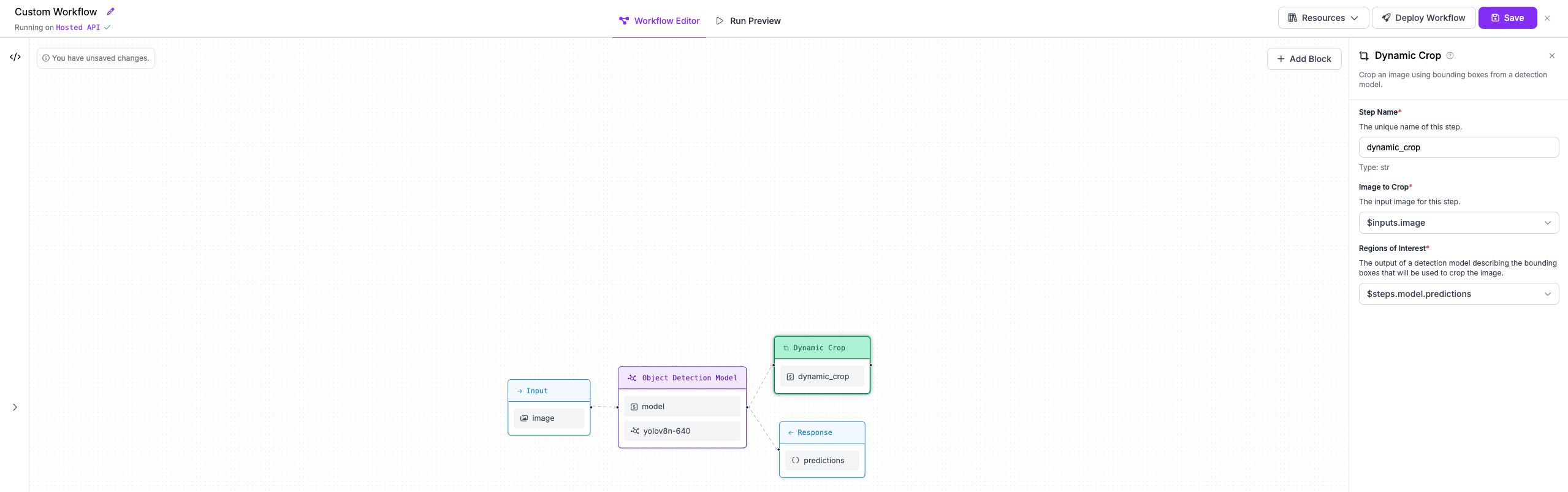
+Next, we are going to add a block to our Workflow that crops the objects that our first model detects.
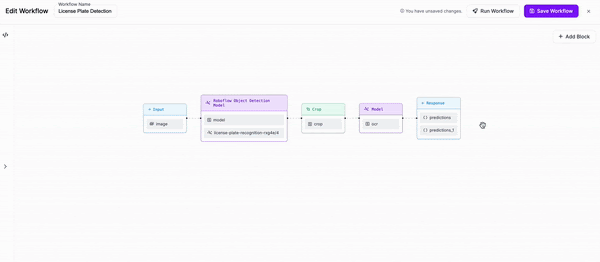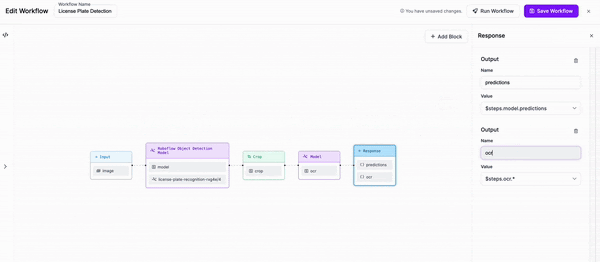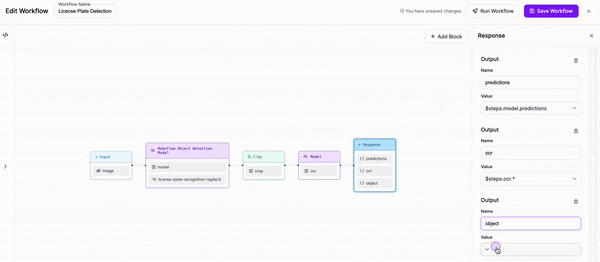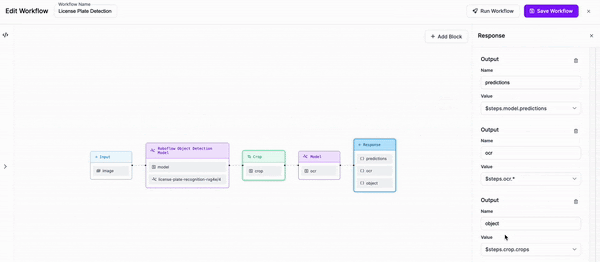
-Finally, we are going to add an output to our response which includes the object that we cropped, alongside the outputs of both our detection model and our OCR model.
+
-
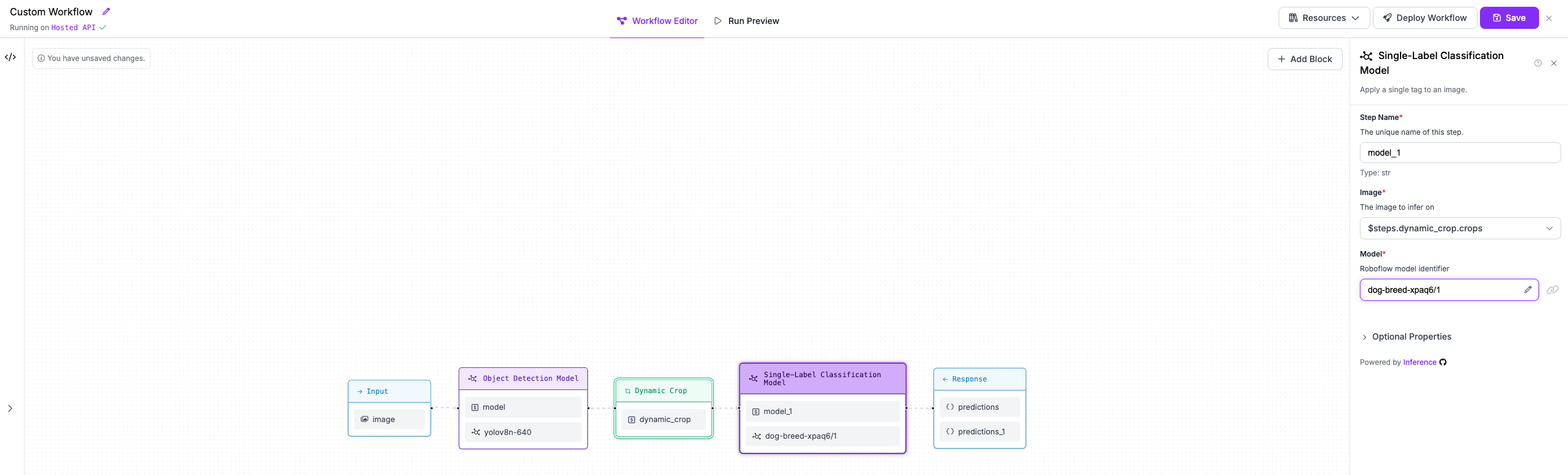
+## Step 4: Classify dog breeds with second stage model
-### Run the Workflow
+We are then going to add a classification model that runs on each crop to classify its content. We will use
+Roboflow Universe model `dog-breed-xpaq6/1`. Please make sure that in the block configuration, the `Image` property
+points to the `crops` output of the Dynamic Crop block.
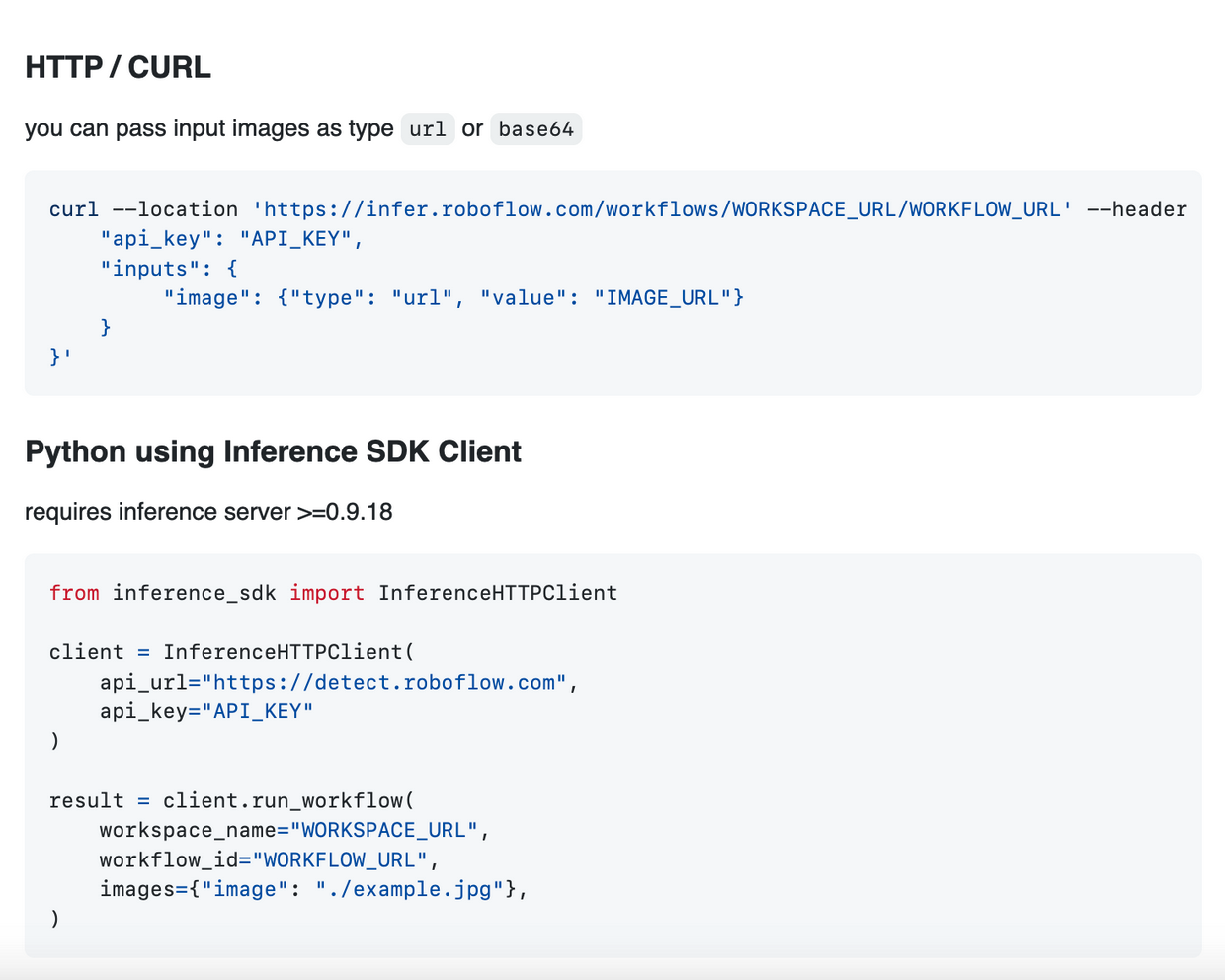
-Selecting the Run Workflow button generates the code snippets to then deploy your Workflow via the Roboflow Hosted API, locally on images via the Inference Server, and locally on video streams via the Inference Pipeline.
-
+
-You now have a workflow you can run on your own hardware!
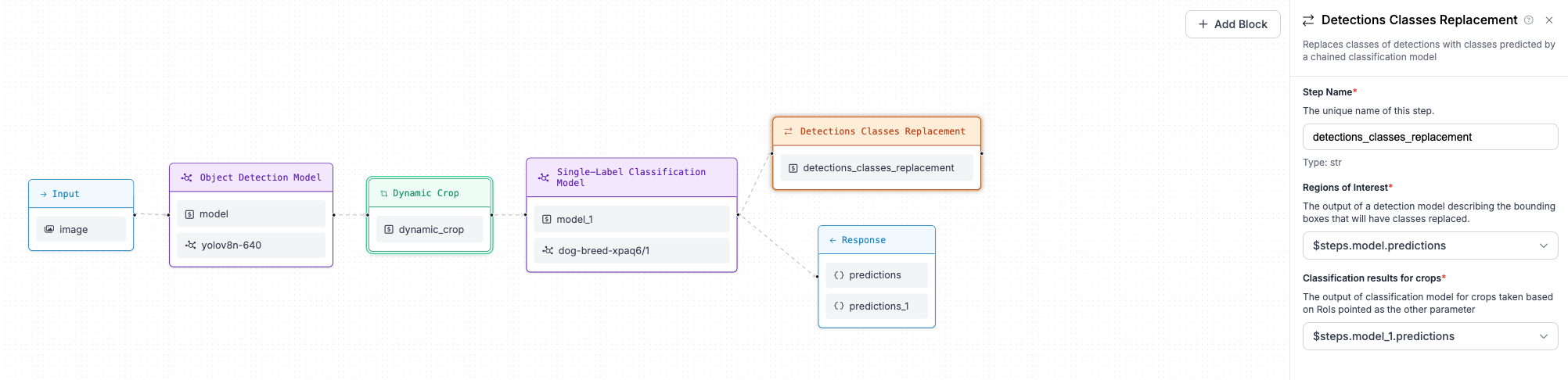
+## Step 5: Replace Bounding Box classes with classification model predictions
-## Example (Code, Advanced)
+When each crop is classified, we would like to assign the class predicted for each crop (dog breed) as a class
+of the dog bounding boxes from the object detection model . To do this we use Detections Classes Replacement block,
+which accepts a reference to predictions of a object detection model, as well as a reference to the classification
+results on the crops.
-Workflows allow you to define multi-step processes that run one or more models and return a result based on the output of the models.
+
-You can create and deploy workflows in the cloud or in Inference.
-To create an advanced workflow for use with Inference, you need to define a specification. A specification is a JSON document that states:
+## Step 6: Visualise predictions
-1. The version of workflows you are using.
-2. The expected inputs.
-3. The steps the workflow should run (i.e. models to run, filters to apply, etc.).
-4. The expected output format.
+As a final step of the workflow, we would like to visualize our predictions. We will use two
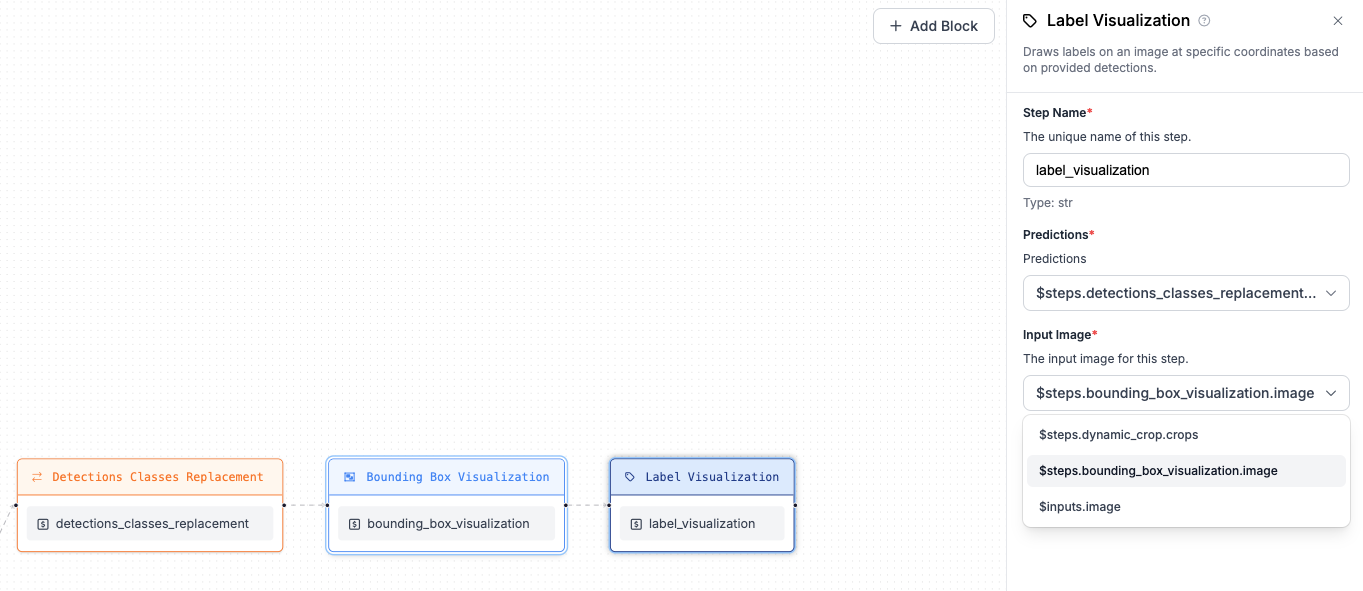
+visualization blocks: Bounding Box Visualization and Label Visualization chained together.
+At first, add Bounding Box Visualization referring to `$inputs.image` for the Image property (that's the
+image sent as your input to workflow), the second step (Label Visualization) however, should point to
+the output of Bounding Box Visualization step. Both visualization steps should refer to predictions
+from the Detections Classes Replacement step.
-In this guide, we walk through how to create a basic workflow that completes three steps:
+
-1. Run a model to detect objects in an image.
-2. Crops each region.
-3. Runs OCR on each region.
+## Step 7: Construct output
+You have everything ready to construct your workflow output. You can use any intermediate step output that you
+need, but in this example we will only select bounding boxes with replaced classes (output from Detections
+Classes Replacement step) and visualisation (output from Label Visualization step).
-You can use the guidance below as a template to learn the structure of workflows, or verbatim to create your own detect-then-OCR workflows.
-## Step #1: Define an Input
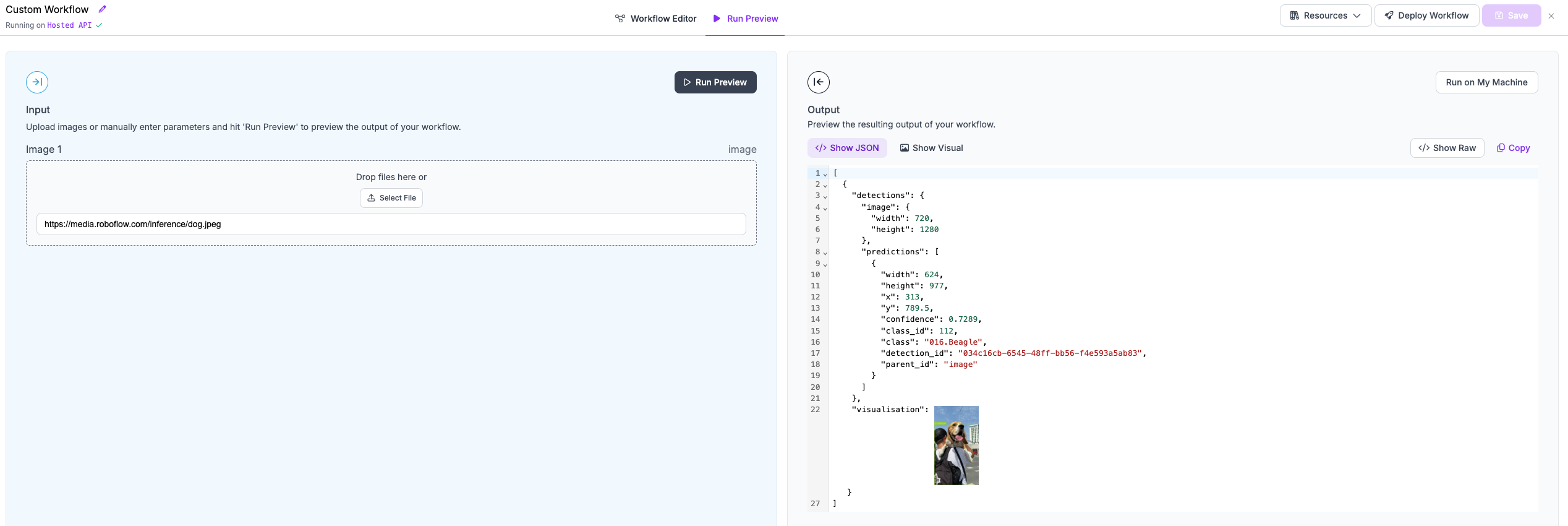
+## Step 8: Running the workflow
+Now your workflow, is ready. You can click the `Save` button and move to the `Run Preview` panel.
-Workflows require a specification to run. A specification takes the follwoing form:
+We will run our workflow against the following example image `https://media.roboflow.com/inference/dog.jpeg`.
+Here are the results
-```python
-SPECIFICATION = {
- "specification": {
- "version": "1.0",
- "inputs": [],
- "steps": [],
- "outputs": []
- }
-}
-```
+
-Within this structure, we need to define our:
+Clicking on the `Show Visual` button you will find results of our visualization efforts.
+
-1. Model inputs
-2. The steps to run
-3. The expected output
-First, let's define our inputs.
+## Different ways of running your workflow
+Your workflow is now saved on the Roboflow Platform. This means you can run it in multiple different ways, including:
-For this workflow, we will specify an image input:
+- HTTP request to Roboflow Hosted API
-```json
-"steps": [
- { "type": "InferenceImage", "name": "image" }, # definition of input image
-]
-```
+- HTTP request to your local instance of `inference server`
-## Step #2: Define Processing Steps
+- on video
-Next, we need to define our processing steps. For this guide, we want to:
+To see code snippets, click the `Deploy Workflow` button:
+
-1. Run a model to detect license plates.
-2. Crop each license plate.
-3. Run OCR on each license plate.
+## Workflow definition for quick reproduction
-We can define these steps as follows:
+To make it easier to reproduce the workflow, below you can find a workflow definition you can copy-paste to UI editor.
-```json
-"steps": [
+??? Tip "Workflow definition"
+
+ ```json
{
- "type": "ObjectDetectionModel", # definition of object detection model
- "name": "plates_detector",
- "image": "$inputs.image", # linking input image into detection model
- "model_id": "vehicle-registration-plates-trudk/2", # pointing model to be used
- },
+ "version": "1.0",
+ "inputs": [
{
- "type": "DetectionOffset", # DocTR model usually works better if there is slight padding around text to be detected - hence we are offseting predictions
- "name": "offset",
- "predictions": "$steps.plates_detector.predictions", # reference to the object detection model output
- "offset_x": 200, # value of offset
- "offset_y": 40, # value of offset
- },
- {
- "type": "Crop", # we would like to run OCR against each and every plate detected - hece we are cropping inputr image using offseted predictions
- "name": "cropping",
- "image": "$inputs.image", # we need to point image to crop
- "detections": "$steps.offset.predictions", # we need to point detections that will be used to crop image (in this case - we use offseted prediction)
- },
- {
- "type": "OCRModel", # we define OCR model
- "name": "step_ocr",
- "image": "$steps.cropping.crops", # OCR model as an input takes a reference to crops that were created based on detections
- },
-],
-```
-
-## Step #3: Define an Output
-
-Finally, we need to define the output for our workflow:
-
-```json
-"outputs": [
- { "type": "JsonField", "name": "predictions", "selector": "$steps.plates_detector.predictions" }, # output with object detection model predictions
- { "type": "JsonField", "name": "image", "selector": "$steps.plates_detector.image" }, # output with image metadata - required by `supervision`
- { "type": "JsonField", "name": "recognised_plates", "selector": "$steps.step_ocr.result" }, # field that will retrieve OCR result
- { "type": "JsonField", "name": "crops", "selector": "$steps.cropping.crops" }, # crops that were made based on plates detections - used here just to ease visualisation
-]
-```
-
-## Step #4: Run Your Workflow
-
-Now that we have our specification, we can run our workflow using the Inference SDK.
-
-=== "Run Locally with Inference"
-
- Use `inference_cli` to start server
-
- ```bash
- inference server start
- ```
-
- ```python
- from inference_sdk import InferenceHTTPClient, VisualisationResponseFormat, InferenceConfiguration
- import supervision as sv
- import cv2
- from matplotlib import pyplot as plt
-
- client = InferenceHTTPClient(
- api_url="http://127.0.0.1:9001",
- api_key="YOUR_API_KEY"
- )
-
- client.configure(
- InferenceConfiguration(output_visualisation_format=VisualisationResponseFormat.NUMPY)
- )
-
- license_plate_image_1 = cv2.imread("./images/license_plate_1.jpg")
-
- license_plate_result_1 = client.infer_from_workflow(
- specification=READING_PLATES_SPECIFICATION["specification"],
- images={"image": license_plate_image_1},
- )
-
- plt.title(f"Recognised plate: {license_plate_result_1['recognised_plates']}")
- plt.imshow(license_plate_result_1["crops"][0]["value"][:, :, ::-1])
- plt.show()
- ```
-
- Here are the results:
-
- 
-
-=== "Run in the Roboflow Cloud"
-
- ```python
- from inference_sdk import InferenceHTTPClient
-
- client = InferenceHTTPClient(
- api_url="https://detect.roboflow.com",
- api_key="YOUR_API_KEY"
- )
-
- client.configure(
- InferenceConfiguration(output_visualisation_format=VisualisationResponseFormat.NUMPY)
- )
-
- license_plate_image_1 = cv2.imread("./images/license_plate_1.jpg")
-
- license_plate_result_1 = client.infer_from_workflow(
- specification=READING_PLATES_SPECIFICATION["specification"],
- images={"image": license_plate_image_1},
- )
-
- plt.title(f"Recognised plate: {license_plate_result_1['recognised_plates']}")
- plt.imshow(license_plate_result_1["crops"][0]["value"][:, :, ::-1])
- plt.show()
+ "type": "InferenceImage",
+ "name": "image"
+ }
+ ],
+ "steps": [
+ {
+ "type": "roboflow_core/roboflow_object_detection_model@v1",
+ "name": "model",
+ "images": "$inputs.image",
+ "model_id": "yolov8n-640"
+ },
+ {
+ "type": "roboflow_core/dynamic_crop@v1",
+ "name": "dynamic_crop",
+ "images": "$inputs.image",
+ "predictions": "$steps.model.predictions"
+ },
+ {
+ "type": "roboflow_core/roboflow_classification_model@v1",
+ "name": "model_1",
+ "images": "$steps.dynamic_crop.crops",
+ "model_id": "dog-breed-xpaq6/1"
+ },
+ {
+ "type": "roboflow_core/detections_classes_replacement@v1",
+ "name": "detections_classes_replacement",
+ "object_detection_predictions": "$steps.model.predictions",
+ "classification_predictions": "$steps.model_1.predictions"
+ },
+ {
+ "type": "roboflow_core/bounding_box_visualization@v1",
+ "name": "bounding_box_visualization",
+ "predictions": "$steps.detections_classes_replacement.predictions",
+ "image": "$inputs.image"
+ },
+ {
+ "type": "roboflow_core/label_visualization@v1",
+ "name": "label_visualization",
+ "predictions": "$steps.detections_classes_replacement.predictions",
+ "image": "$steps.bounding_box_visualization.image"
+ }
+ ],
+ "outputs": [
+ {
+ "type": "JsonField",
+ "name": "detections",
+ "coordinates_system": "own",
+ "selector": "$steps.detections_classes_replacement.predictions"
+ },
+ {
+ "type": "JsonField",
+ "name": "visualisation",
+ "coordinates_system": "own",
+ "selector": "$steps.label_visualization.image"
+ }
+ ]
+ }
```
- Here are the results:
-
- 
## Next Steps
Now that you have created and run your first workflow, you can explore our other supported blocks and create a more complex workflow.
Refer to our [Supported Blocks](/workflows/blocks/) documentation to learn more about what blocks are supported.
+We also recommend reading the [Understanding workflows](/workflows/understanding/) page.
diff --git a/docs/workflows/create_workflow_block.md b/docs/workflows/create_workflow_block.md
new file mode 100644
index 0000000000..ae2dab937f
--- /dev/null
+++ b/docs/workflows/create_workflow_block.md
@@ -0,0 +1,1913 @@
+# Creating Workflow blocks
+
+Workflows blocks development requires an understanding of the
+Workflow Ecosystem. Before diving deeper into the details, let's summarize the
+required knowledge:
+
+Understanding of [Workflow execution](/workflows/workflow_execution/), in particular:
+
+* what is the relation of Workflow blocks and steps in Workflow definition
+
+* how Workflow blocks and their manifests are used by [Workflows Compiler](/workflows/workflows_compiler/)
+
+* what is the `dimensionality level` of batch-oriented data passing through Workflow
+
+* how [Execution Engine](/workflows/workflows_execution_engine/) interacts with step, regarding
+its inputs and outputs
+
+* what is the nature and role of [Workflow `kinds`](/workflows/kinds/)
+
+* understanding how [`pydantic`](https://docs.pydantic.dev/latest/) works
+
+## Prototypes
+
+To create a Workflow block you need some amount of imports from the core of Workflows library.
+Here is the list of imports that you may find useful while creating a block:
+
+```python
+from inference.core.workflows.execution_engine.entities.base import (
+ Batch, # batches of data will come in Batch[X] containers
+ OutputDefinition, # class used to declare outputs in your manifest
+ WorkflowImageData, # internal representation of image
+ # - use whenever your input kind is image
+)
+
+from inference.core.workflows.prototypes.block import (
+ BlockResult, # type alias for result of `run(...)` method
+ WorkflowBlock, # base class for your block
+ WorkflowBlockManifest, # base class for block manifest
+)
+
+from inference.core.workflows.execution_engine.entities.types import *
+# module with `kinds` from the core library
+```
+
+The most important are:
+
+* `WorkflowBlock` - base class for your block
+
+* `WorkflowBlockManifest` - base class for block manifest
+
+## Block manifest
+
+A manifest is a crucial component of a Workflow block that defines a prototype
+for step declaration that can be placed in a Workflow definition to use the block.
+In particular, it:
+
+* **Uses `pydantic` to power syntax parsing of Workflows definitions:**
+It inherits from [`pydantic BaseModel`](https://docs.pydantic.dev/latest/api/base_model/) features to parse and
+validate Workflow definitions. This schema can also be automatically exported to a format compatible with the
+Workflows UI, thanks to `pydantic's` integration with the OpenAPI standard.
+
+* **Defines Data Bindings:** It specifies which fields in the manifest are selectors for data flowing through
+the workflow during execution and indicates their kinds.
+
+* **Describes Block Outputs:** It outlines the outputs that the block will produce.
+
+* **Specifies Dimensionality:** It details the properties related to input and output dimensionality.
+
+* **Indicates Batch Inputs and Empty Values:** It informs the Execution Engine whether the step accepts batch
+inputs and empty values.
+
+* **Ensures Compatibility:** It dictates the compatibility with different Execution Engine versions to maintain
+stability. For more details, see [versioning](/workflows/versioning/).
+
+### Scaffolding for manifest
+
+To understand how manifests work, let's define one step-by-step. The example block that we build here will be
+calculating images similarity. We start from imports and class scaffolding:
+
+```python
+from typing import Literal
+from inference.core.workflows.prototypes.block import (
+ WorkflowBlockManifest,
+)
+
+class ImagesSimilarityManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/images_similarity@v1"]
+ name: str
+```
+
+This is the minimal representation of a manifest. It defines two special fields that are important for
+Compiler and Execution engine:
+
+* `type` - required to parse syntax of Workflows definitions based on dynamic pool of blocks - this is the
+[`pydantic` type discriminator](https://docs.pydantic.dev/latest/concepts/unions/#discriminated-unions) that lets the Compiler understand which block manifest is to be verified when
+parsing specific steps in a Workflow definition
+
+* `name` - this property will be used to give the step a unique name and let other steps selects it via selectors
+
+### Adding batch-oriented inputs
+
+We want our step to take two batch-oriented inputs with images to be compared - so effectively
+we will be creating SIMD block.
+
+??? example "Adding batch-oriented inputs"
+
+ Let's see how to add definitions of those inputs to manifest:
+
+ ```{ .py linenums="1" hl_lines="2 6 7 8 9 17 18 19 20 21 22"}
+ from typing import Literal, Union
+ from pydantic import Field
+ from inference.core.workflows.prototypes.block import (
+ WorkflowBlockManifest,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ StepOutputImageSelector,
+ WorkflowImageSelector,
+ )
+
+ class ImagesSimilarityManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/images_similarity@v1"]
+ name: str
+ # all properties apart from `type` and `name` are treated as either
+ # definitions of batch-oriented data to be processed by block or its
+ # parameters that influence execution of steps created based on block
+ image_1: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="First image to calculate similarity",
+ )
+ image_2: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="Second image to calculate similarity",
+ )
+ ```
+
+ * in the lines `2-9`, we've added a couple of imports to ensure that we have everything needed
+
+ * line `17` defines `image_1` parameter - as manifest is prototype for Workflow Definition,
+ the only way to tell about image to be used by step is to provide selector - we have
+ two specialised types in core library that can be used - `WorkflowImageSelector` and `StepOutputImageSelector`.
+ If you look deeper into codebase, you will discover those are type aliases - telling `pydantic`
+ to expect string matching `$inputs.{name}` and `$steps.{name}.*` patterns respectively, additionally providing
+ extra schema field metadata that tells Workflows ecosystem components that the `kind` of data behind selector is
+ [image](/workflows/kinds/batch_image/).
+
+ * denoting `pydantic` `Field(...)` attribute in the last parts of line `17` is optional, yet appreciated,
+ especially for blocks intended to cooperate with Workflows UI
+
+ * starting in line `20`, you can find definition of `image_2` parameter which is very similar to `image_1`.
+
+
+Such definition of manifest can handle the following step declaration in Workflow definition:
+
+```json
+{
+ "type": "my_plugin/images_similarity@v1",
+ "name": "my_step",
+ "image_1": "$inputs.my_image",
+ "image_2": "$steps.image_transformation.image"
+}
+```
+
+This definition will make the Compiler and Execution Engine:
+
+* select as a step prototype the block which declared manifest with type discriminator being
+`my_plugin/images_similarity@v1`
+
+* supply two parameters for the steps run method:
+
+ * `input_1` of type `WorkflowImageData` which will be filled with image submitted as Workflow execution input
+
+ * `imput_2` of type `WorkflowImageData` which will be generated at runtime, by another step called
+ `image_transformation`
+
+
+### Adding parameter to the manifest
+
+Let's now add the parameter that will influence step execution. The parameter is not assumed to be
+batch-oriented and will affect all batch elements passed to the step.
+
+??? example "Adding parameter to the manifest"
+
+ ```{ .py linenums="1" hl_lines="9 10 11 26 27 28 29 30 31 32"}
+ from typing import Literal, Union
+ from pydantic import Field
+ from inference.core.workflows.prototypes.block import (
+ WorkflowBlockManifest,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ StepOutputImageSelector,
+ WorkflowImageSelector,
+ FloatZeroToOne,
+ WorkflowParameterSelector,
+ FLOAT_ZERO_TO_ONE_KIND,
+ )
+
+ class ImagesSimilarityManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/images_similarity@v1"]
+ name: str
+ # all properties apart from `type` and `name` are treated as either
+ # definitions of batch-oriented data to be processed by block or its
+ # parameters that influence execution of steps created based on block
+ image_1: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="First image to calculate similarity",
+ )
+ image_2: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="Second image to calculate similarity",
+ )
+ similarity_threshold: Union[
+ FloatZeroToOne,
+ WorkflowParameterSelector(kind=[FLOAT_ZERO_TO_ONE_KIND]),
+ ] = Field(
+ default=0.4,
+ description="Threshold to assume that images are similar",
+ )
+ ```
+
+ * line `9` imports `FloatZeroToOne` which is type alias providing validation
+ for float values in range 0.0-1.0 - this is based on native `pydantic` mechanism and
+ everyone could create this type annotation locally in module hosting block
+
+ * line `10` imports function `WorkflowParameterSelector(...)` capable to dynamically create
+ `pydantic` type annotation for selector to workflow input parameter (matching format `$inputs.param_name`),
+ declaring union of kinds compatible with the field
+
+ * line `11` imports [`float_zero_to_one`](/workflows/kinds/float_zero_to_one) `kind` definition which will be used later
+
+ * in line `26` we start defining parameter called `similarity_threshold`. Manifest will accept
+ either float values (in range `[0.0-1.0]`) or selector to workflow input of `kind`
+ [`float_zero_to_one`](/workflows/kinds/float_zero_to_one). Please point out on how
+ function creating type annotation (`WorkflowParameterSelector(...)`) is used -
+ in particular, expected `kind` of data is passed as list of `kinds` - representing union
+ of expected data `kinds`.
+
+Such definition of manifest can handle the following step declaration in Workflow definition:
+
+```{ .json linenums="1" hl_lines="6"}
+{
+ "type": "my_plugin/images_similarity@v1",
+ "name": "my_step",
+ "image_1": "$inputs.my_image",
+ "image_2": "$steps.image_transformation.image",
+ "similarity_threshold": "$inputs.my_similarity_threshold"
+}
+```
+
+or alternatively:
+
+```{ .json linenums="1" hl_lines="6"}
+{
+ "type": "my_plugin/images_similarity@v1",
+ "name": "my_step",
+ "image_1": "$inputs.my_image",
+ "image_2": "$steps.image_transformation.image",
+ "similarity_threshold": "0.5"
+}
+```
+
+??? hint "LEARN MORE: Selecting step outputs"
+
+ Our siplified example showcased declaration of properties that accept selectors to
+ images produced by other steps via `StepOutputImageSelector`.
+
+ You can use function `StepOutputSelector(...)` creating field annotations dynamically
+ to express the that block accepts batch-oriented outputs from other steps of specified
+ kinds
+
+ ```{ .py linenums="1" hl_lines="9 10 25"}
+ from typing import Literal, Union
+ from pydantic import Field
+ from inference.core.workflows.prototypes.block import (
+ WorkflowBlockManifest,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ StepOutputImageSelector,
+ WorkflowImageSelector,
+ StepOutputSelector,
+ NUMPY_ARRAY_KIND,
+ )
+
+ class ImagesSimilarityManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/images_similarity@v1"]
+ name: str
+ # all properties apart from `type` and `name` are treated as either
+ # definitions of batch-oriented data to be processed by block or its
+ # parameters that influence execution of steps created based on block
+ image_1: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="First image to calculate similarity",
+ )
+ image_2: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="Second image to calculate similarity",
+ )
+ example: StepOutputSelector(kind=[NUMPY_ARRAY_KIND])
+ ```
+
+### Declaring block outputs
+
+Our manifest is ready regarding properties that can be declared in Workflow definitions,
+but we still need to provide additional information for the Execution Engine to successfully
+run the block.
+
+??? example "Declaring block outputs"
+
+ Minimal set of information required is outputs description. Additionally,
+ to increase block stability, we advise to provide information about execution engine
+ compatibility.
+
+ ```{ .py linenums="1" hl_lines="1 5 13 33-40 42-44"}
+ from typing import Literal, Union, List, Optional
+ from pydantic import Field
+ from inference.core.workflows.prototypes.block import (
+ WorkflowBlockManifest,
+ OutputDefinition,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ StepOutputImageSelector,
+ WorkflowImageSelector,
+ FloatZeroToOne,
+ WorkflowParameterSelector,
+ FLOAT_ZERO_TO_ONE_KIND,
+ BOOLEAN_KIND,
+ )
+
+ class ImagesSimilarityManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/images_similarity@v1"]
+ name: str
+ image_1: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="First image to calculate similarity",
+ )
+ image_2: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="Second image to calculate similarity",
+ )
+ similarity_threshold: Union[
+ FloatZeroToOne,
+ WorkflowParameterSelector(kind=[FLOAT_ZERO_TO_ONE_KIND]),
+ ] = Field(
+ default=0.4,
+ description="Threshold to assume that images are similar",
+ )
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [
+ OutputDefinition(
+ name="images_match",
+ kind=[BOOLEAN_KIND],
+ )
+ ]
+
+ @classmethod
+ def get_execution_engine_compatibility(cls) -> Optional[str]:
+ return ">=1.0.0,<2.0.0"
+ ```
+
+ * line `1` contains additional imports from `typing`
+
+ * line `5` imports class that is used to describe step outputs
+
+ * line `13` imports [`boolean`](/workflows/kinds/boolean) `kind` to be used
+ in outputs definitions
+
+ * lines `33-40` declare class method to specify outputs from the block -
+ each entry in list declare one return property for each batch element and its `kind`.
+ Our block will return boolean flag `images_match` for each pair of images.
+
+ * lines `42-44` declare compatibility of the block with Execution Engine -
+ see [versioning page](/workflows/versioning/) for more details
+
+As a result of those changes:
+
+* Execution Engine would understand that steps created based on this block
+are supposed to deliver specified outputs and other steps can refer to those outputs
+in their inputs
+
+* the blocks loading mechanism will not load the block given that Execution Engine is not in version `v1`
+
+??? hint "LEARN MORE: Dynamic outputs"
+
+ Some blocks may not be able to arbitrailry define their outputs using
+ classmethod - regardless of the content of step manifest that is available after
+ parsing. To support this we introduced the following convention:
+
+ * classmethod `describe_outputs(...)` shall return list with one element of
+ name `*` and kind `*` (aka `WILDCARD_KIND`)
+
+ * additionally, block manifest should implement instance method `get_actual_outputs(...)`
+ that provides list of actual outputs that can be generated based on filled manifest data
+
+ ```{ .py linenums="1" hl_lines="14 35-42 44-49"}
+ from typing import Literal, Union, List, Optional
+ from pydantic import Field
+ from inference.core.workflows.prototypes.block import (
+ WorkflowBlockManifest,
+ OutputDefinition,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ StepOutputImageSelector,
+ WorkflowImageSelector,
+ FloatZeroToOne,
+ WorkflowParameterSelector,
+ FLOAT_ZERO_TO_ONE_KIND,
+ BOOLEAN_KIND,
+ WILDCARD_KIND,
+ )
+
+ class ImagesSimilarityManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/images_similarity@v1"]
+ name: str
+ image_1: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="First image to calculate similarity",
+ )
+ image_2: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="Second image to calculate similarity",
+ )
+ similarity_threshold: Union[
+ FloatZeroToOne,
+ WorkflowParameterSelector(kind=[FLOAT_ZERO_TO_ONE_KIND]),
+ ] = Field(
+ default=0.4,
+ description="Threshold to assume that images are similar",
+ )
+ outputs: List[str]
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [
+ OutputDefinition(
+ name="*",
+ kind=[WILDCARD_KIND],
+ ),
+ ]
+
+ def get_actual_outputs(self) -> List[OutputDefinition]:
+ # here you have access to `self`:
+ return [
+ OutputDefinition(name=e, kind=[BOOLEAN_KIND])
+ for e in self.outputs
+ ]
+ ```
+
+
+## Definition of block class
+
+At this stage, the manifest of our simple block is ready, we will continue
+with our example. You can check out the [advanced topics](#advanced-topics) section for more details that would just
+be a distractions now.
+
+### Base implementation
+
+Having the manifest ready, we can prepare baseline implementation of the
+block.
+
+??? example "Block scaffolding"
+
+ ```{ .py linenums="1" hl_lines="1 5 6 8-11 56-68"}
+ from typing import Literal, Union, List, Optional, Type
+ from pydantic import Field
+ from inference.core.workflows.prototypes.block import (
+ WorkflowBlockManifest,
+ WorkflowBlock,
+ BlockResult,
+ )
+ from inference.core.workflows.execution_engine.entities.base import (
+ OutputDefinition,
+ WorkflowImageData,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ StepOutputImageSelector,
+ WorkflowImageSelector,
+ FloatZeroToOne,
+ WorkflowParameterSelector,
+ FLOAT_ZERO_TO_ONE_KIND,
+ BOOLEAN_KIND,
+ )
+
+ class ImagesSimilarityManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/images_similarity@v1"]
+ name: str
+ image_1: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="First image to calculate similarity",
+ )
+ image_2: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="Second image to calculate similarity",
+ )
+ similarity_threshold: Union[
+ FloatZeroToOne,
+ WorkflowParameterSelector(kind=[FLOAT_ZERO_TO_ONE_KIND]),
+ ] = Field(
+ default=0.4,
+ description="Threshold to assume that images are similar",
+ )
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [
+ OutputDefinition(
+ name="images_match",
+ kind=[BOOLEAN_KIND],
+ ),
+ ]
+
+ @classmethod
+ def get_execution_engine_compatibility(cls) -> Optional[str]:
+ return ">=1.0.0,<2.0.0"
+
+
+ class ImagesSimilarityBlock(WorkflowBlock):
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return ImagesSimilarityManifest
+
+ def run(
+ self,
+ image_1: WorkflowImageData,
+ image_2: WorkflowImageData,
+ similarity_threshold: float,
+ ) -> BlockResult:
+ pass
+ ```
+
+ * lines `1`, `5-6` and `8-9` added changes into import surtucture to
+ provide additional symbols required to properly define block class and all
+ of its methods signatures
+
+ * line `59` defines class method `get_manifest(...)` to simply return
+ the manifest class we cretaed earlier
+
+ * lines `62-68` define `run(...)` function, which Execution Engine
+ will invoke with data to get desired results
+
+### Providing implementation for block logic
+
+Let's now add an example implementation of the `run(...)` method to our block, such that
+it can produce meaningful results.
+
+!!! Note
+
+ The Content of this section is supposed to provide examples on how to interact
+ with the Workflow ecosystem as block creator, rather than providing robust
+ implementation of the block.
+
+??? example "Implementation of `run(...)` method"
+
+ ```{ .py linenums="1" hl_lines="3 56-58 70-81"}
+ from typing import Literal, Union, List, Optional, Type
+ from pydantic import Field
+ import cv2
+
+ from inference.core.workflows.prototypes.block import (
+ WorkflowBlockManifest,
+ WorkflowBlock,
+ BlockResult,
+ )
+ from inference.core.workflows.execution_engine.entities.base import (
+ OutputDefinition,
+ WorkflowImageData,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ StepOutputImageSelector,
+ WorkflowImageSelector,
+ FloatZeroToOne,
+ WorkflowParameterSelector,
+ FLOAT_ZERO_TO_ONE_KIND,
+ BOOLEAN_KIND,
+ )
+
+ class ImagesSimilarityManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/images_similarity@v1"]
+ name: str
+ image_1: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="First image to calculate similarity",
+ )
+ image_2: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="Second image to calculate similarity",
+ )
+ similarity_threshold: Union[
+ FloatZeroToOne,
+ WorkflowParameterSelector(kind=[FLOAT_ZERO_TO_ONE_KIND]),
+ ] = Field(
+ default=0.4,
+ description="Threshold to assume that images are similar",
+ )
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [
+ OutputDefinition(
+ name="images_match",
+ kind=[BOOLEAN_KIND],
+ ),
+ ]
+
+ @classmethod
+ def get_execution_engine_compatibility(cls) -> Optional[str]:
+ return ">=1.0.0,<2.0.0"
+
+
+ class ImagesSimilarityBlock(WorkflowBlock):
+
+ def __init__(self):
+ self._sift = cv2.SIFT_create()
+ self._matcher = cv2.FlannBasedMatcher(dict(algorithm=1, trees=5), dict(checks=50))
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return ImagesSimilarityManifest
+
+ def run(
+ self,
+ image_1: WorkflowImageData,
+ image_2: WorkflowImageData,
+ similarity_threshold: float,
+ ) -> BlockResult:
+ image_1_gray = cv2.cvtColor(image_1.numpy_image, cv2.COLOR_BGR2GRAY)
+ image_2_gray = cv2.cvtColor(image_2.numpy_image, cv2.COLOR_BGR2GRAY)
+ kp_1, des_1 = self._sift.detectAndCompute(image_1_gray, None)
+ kp_2, des_2 = self._sift.detectAndCompute(image_2_gray, None)
+ matches = self._matcher.knnMatch(des_1, des_2, k=2)
+ good_matches = []
+ for m, n in matches:
+ if m.distance < similarity_threshold * n.distance:
+ good_matches.append(m)
+ return {
+ "images_match": len(good_matches) > 0,
+ }
+ ```
+
+ * in line `3` we import OpenCV
+
+ * lines `56-58` defines block constructor, thanks to this - state of block
+ is initialised once and live through consecutive invocation of `run(...)` method - for
+ instance when Execution Engine runs on consecutive frames of video
+
+ * lines `70-81` provide implementation of block functionality - the details are trully not
+ important regarding Workflows ecosystem, but there are few details you should focus:
+
+ * lines `70` and `71` make use of `WorkflowImageData` abstraction, showcasing how
+ `numpy_image` property can be used to get `np.ndarray` from internal representation of images
+ in Workflows. We advise to expole remaining properties of `WorkflowImageData` to discover more.
+
+ * result of workflow block execution, declared in lines `79-81` is in our case just a dictionary
+ **with the keys being the names of outputs declared in manifest**, in line `44`. Be sure to provide all
+ declared outputs - otherwise Execution Engine will raise error.
+
+You may ask yourself how it is possible that implemented block accepts batch-oriented workflow input, but do not
+operate on batches directly. This is due to the fact that the default block behaviour is to run one-by-one against
+all elements of input batches. We will show how to change that in [advanced topics](#advanced-topics) section.
+
+!!! note
+
+ One important note: blocks, like all other classes, have constructors that may initialize a state. This state can
+ persist across multiple Workflow runs when using the same instance of the Execution Engine. If the state management
+ needs to be aware of which batch element it processes (e.g., in object tracking scenarios), the block creator
+ should use dedicated batch-oriented inputs. These inputs, provide relevant metadatadata — like the
+ `WorkflowVideoMetadata` input, which is crucial for tracking use cases and can be used along with `WorkflowImage`
+ input in a block implementing tracker.
+
+ The ecosystem is evolving, and new input types will be introduced over time. If a specific input type needed for
+ a use case is not available, an alternative is to design the block to process entire input batches. This way,
+ you can rely on the Batch container's indices property, which provides an index for each batch element, allowing
+ you to maintain the correct order of processing.
+
+
+## Exposing block in `plugin`
+
+Now, your block is ready to be used, but if you declared step using it in your Workflow definition you
+would see an error. This is because no plugin exports the block you just created. Details of blocks bundling
+will be covered in [separate page](/workflows/blocks_bundling/), but the remaining thing to do is to
+add block class into list returned from your plugins' `load_blocks(...)` function:
+
+```python
+# __init__.py of your plugin
+
+from my_plugin.images_similarity.v1 import ImagesSimilarityBlock
+# this is example import! requires adjustment
+
+def load_blocks():
+ return [ImagesSimilarityBlock]
+```
+
+
+## Advanced topics
+
+### Blocks processing batches of inputs
+
+Sometimes, performance of your block may benefit if all input data is processed at once as batch. This may
+happen for models running on GPU. Such mode of operation is supported for Workflows blocks - here is the example
+on how to use it for your block.
+
+??? example "Implementation of blocks accepting batches"
+
+ ```{ .py linenums="1" hl_lines="13 41-43 71-72 75-78 86-87"}
+ from typing import Literal, Union, List, Optional, Type
+ from pydantic import Field
+ import cv2
+
+ from inference.core.workflows.prototypes.block import (
+ WorkflowBlockManifest,
+ WorkflowBlock,
+ BlockResult,
+ )
+ from inference.core.workflows.execution_engine.entities.base import (
+ OutputDefinition,
+ WorkflowImageData,
+ Batch,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ StepOutputImageSelector,
+ WorkflowImageSelector,
+ FloatZeroToOne,
+ WorkflowParameterSelector,
+ FLOAT_ZERO_TO_ONE_KIND,
+ BOOLEAN_KIND,
+ )
+
+ class ImagesSimilarityManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/images_similarity@v1"]
+ name: str
+ image_1: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="First image to calculate similarity",
+ )
+ image_2: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
+ description="Second image to calculate similarity",
+ )
+ similarity_threshold: Union[
+ FloatZeroToOne,
+ WorkflowParameterSelector(kind=[FLOAT_ZERO_TO_ONE_KIND]),
+ ] = Field(
+ default=0.4,
+ description="Threshold to assume that images are similar",
+ )
+
+ @classmethod
+ def accepts_batch_input(cls) -> bool:
+ return True
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [
+ OutputDefinition(
+ name="images_match",
+ kind=[BOOLEAN_KIND],
+ ),
+ ]
+
+ @classmethod
+ def get_execution_engine_compatibility(cls) -> Optional[str]:
+ return ">=1.0.0,<2.0.0"
+
+
+ class ImagesSimilarityBlock(WorkflowBlock):
+
+ def __init__(self):
+ self._sift = cv2.SIFT_create()
+ self._matcher = cv2.FlannBasedMatcher(dict(algorithm=1, trees=5), dict(checks=50))
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return ImagesSimilarityManifest
+
+ def run(
+ self,
+ image_1: Batch[WorkflowImageData],
+ image_2: Batch[WorkflowImageData],
+ similarity_threshold: float,
+ ) -> BlockResult:
+ results = []
+ for image_1_element, image_2_element in zip(image_1, image_2):
+ image_1_gray = cv2.cvtColor(image_1_element.numpy_image, cv2.COLOR_BGR2GRAY)
+ image_2_gray = cv2.cvtColor(image_2_element.numpy_image, cv2.COLOR_BGR2GRAY)
+ kp_1, des_1 = self._sift.detectAndCompute(image_1_gray, None)
+ kp_2, des_2 = self._sift.detectAndCompute(image_2_gray, None)
+ matches = self._matcher.knnMatch(des_1, des_2, k=2)
+ good_matches = []
+ for m, n in matches:
+ if m.distance < similarity_threshold * n.distance:
+ good_matches.append(m)
+ results.append({"images_match": len(good_matches) > 0})
+ return results
+ ```
+
+ * line `13` imports `Batch` from core of workflows library - this class represent container which is
+ veri similar to list (but read-only) to keep batch elements
+
+ * lines `41-43` define class method that changes default behaviour of the block and make it capable
+ to process batches
+
+ * changes introduced above made the signature of `run(...)` method to change, now `image_1` and `image_2`
+ are not instances of `WorkflowImageData`, but rather batches of elements of this type
+
+ * lines `75-78`, `86-87` present changes that needed to be introduced to run processing across all batch
+ elements - showcasing how to iterate over batch elements if needed
+
+ * it is important to note how outputs are constructed in line `86` - each element of batch will be given
+ its entry in the list which is returned from `run(...)` method. Order must be aligned with order of batch
+ elements. Each output dictionary must provide all keys declared in block outputs.
+
+### Implementation of flow-control block
+
+Flow-control blocks differs quite substantially from other blocks that just process the data. Here we will show
+how to create a flow control block, but first - a little bit of theory:
+
+* flow-control block is the block that declares compatibility with step selectors in their manifest (selector to step
+is defined as `$steps.{step_name}` - similar to step output selector, but without specification of output name)
+
+* flow-control blocks cannot register outputs, they are meant to return `FlowControl` objects
+
+* `FlowControl` object specify next steps (from selectors provided in step manifest) that for given
+batch element (SIMD flow-control) or whole workflow execution (non-SIMD flow-control) should pick up next
+
+??? example "Implementation of flow-control - SIMD block"
+
+ Example provides and comments out implementation of random continue block
+
+ ```{ .py linenums="1" hl_lines="10 14 26 28-31 55-56"}
+ from typing import List, Literal, Optional, Type, Union
+ import random
+
+ from pydantic import Field
+ from inference.core.workflows.execution_engine.entities.base import (
+ OutputDefinition,
+ WorkflowImageData,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ StepSelector,
+ WorkflowImageSelector,
+ StepOutputImageSelector,
+ )
+ from inference.core.workflows.execution_engine.v1.entities import FlowControl
+ from inference.core.workflows.prototypes.block import (
+ BlockResult,
+ WorkflowBlock,
+ WorkflowBlockManifest,
+ )
+
+
+
+ class BlockManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/random_continue@v1"]
+ name: str
+ image: Union[WorkflowImageSelector, StepOutputImageSelector] = ImageInputField
+ probability: float
+ next_steps: List[StepSelector] = Field(
+ description="Reference to step which shall be executed if expression evaluates to true",
+ examples=[["$steps.on_true"]],
+ )
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return []
+
+ @classmethod
+ def get_execution_engine_compatibility(cls) -> Optional[str]:
+ return ">=1.0.0,<2.0.0"
+
+
+ class RandomContinueBlockV1(WorkflowBlock):
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return BlockManifest
+
+ def run(
+ self,
+ image: WorkflowImageData,
+ probability: float,
+ next_steps: List[str],
+ ) -> BlockResult:
+ if not next_steps or random.random() > probability:
+ return FlowControl()
+ return FlowControl(context=next_steps)
+ ```
+
+ * line `10` imports type annotation for step selector which will be used to
+ notify Execution Engine that the block controls the flow
+
+ * line `14` imports `FlowControl` class which is the only viable response from
+ flow-control block
+
+ * line `26` specifies `image` which is batch-oriented input making the block SIMD -
+ which means that for each element of images batch, block will make random choice on
+ flow-control - if not that input block would operate in non-SIMD mode
+
+ * line `28` defines list of step selectors **which effectively turns the block into flow-control one**
+
+ * lines `55` and `56` show how to construct output - `FlowControl` object accept context being `None`, `string` or
+ `list of strings` - `None` represent flow termination for the batch element, strings are expected to be selectors
+ for next steps, passed in input.
+
+??? example "Implementation of flow-control non-SIMD block"
+
+ Example provides and comments out implementation of random continue block
+
+ ```{ .py linenums="1" hl_lines="9 11 24-27 50-51"}
+ from typing import List, Literal, Optional, Type, Union
+ import random
+
+ from pydantic import Field
+ from inference.core.workflows.execution_engine.entities.base import (
+ OutputDefinition,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ StepSelector,
+ )
+ from inference.core.workflows.execution_engine.v1.entities import FlowControl
+ from inference.core.workflows.prototypes.block import (
+ BlockResult,
+ WorkflowBlock,
+ WorkflowBlockManifest,
+ )
+
+
+
+ class BlockManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/random_continue@v1"]
+ name: str
+ probability: float
+ next_steps: List[StepSelector] = Field(
+ description="Reference to step which shall be executed if expression evaluates to true",
+ examples=[["$steps.on_true"]],
+ )
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return []
+
+ @classmethod
+ def get_execution_engine_compatibility(cls) -> Optional[str]:
+ return ">=1.0.0,<2.0.0"
+
+
+ class RandomContinueBlockV1(WorkflowBlock):
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return BlockManifest
+
+ def run(
+ self,
+ probability: float,
+ next_steps: List[str],
+ ) -> BlockResult:
+ if not next_steps or random.random() > probability:
+ return FlowControl()
+ return FlowControl(context=next_steps)
+ ```
+
+ * line `9` imports type annotation for step selector which will be used to
+ notify Execution Engine that the block controls the flow
+
+ * line `11` imports `FlowControl` class which is the only viable response from
+ flow-control block
+
+ * lines `24-27` defines list of step selectors **which effectively turns the block into flow-control one**
+
+ * lines `50` and `51` show how to construct output - `FlowControl` object accept context being `None`, `string` or
+ `list of strings` - `None` represent flow termination for the batch element, strings are expected to be selectors
+ for next steps, passed in input.
+
+### Nested selectors
+
+Some block will require list of selectors or dictionary of selectors to be
+provided in block manifest field. Version `v1` of Execution Engine supports only
+one level of nesting - so list of lists of selectors or dictionary with list of selectors
+will not be recognised properly.
+
+Practical use cases showcasing usage of nested selectors are presented below.
+
+#### Fusion of predictions from variable number of models
+
+Let's assume that you want to build a block to get majority vote on multiple classifiers predictions - then you would
+like your run method to look like that:
+
+```python
+# pseud-code here
+def run(self, predictions: List[dict]) -> BlockResult:
+ predicted_classes = [p["class"] for p in predictions]
+ counts = Counter(predicted_classes)
+ return {"top_class": counts.most_common(1)[0]}
+```
+
+??? example "Nested selectors - models ensemble"
+
+ ```{ .py linenums="1" hl_lines="23-26 50"}
+ from typing import List, Literal, Optional, Type
+
+ from pydantic import Field
+ import supervision as sv
+ from inference.core.workflows.execution_engine.entities.base import (
+ OutputDefinition,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ StepOutputSelector,
+ BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND,
+ )
+ from inference.core.workflows.prototypes.block import (
+ BlockResult,
+ WorkflowBlock,
+ WorkflowBlockManifest,
+ )
+
+
+
+ class BlockManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/fusion_of_predictions@v1"]
+ name: str
+ predictions: List[StepOutputSelector(kind=[BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND])] = Field(
+ description="Selectors to step outputs",
+ examples=[["$steps.model_1.predictions", "$steps.model_2.predictions"]],
+ )
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [
+ OutputDefinition(
+ name="predictions",
+ kind=[BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND],
+ )
+ ]
+
+ @classmethod
+ def get_execution_engine_compatibility(cls) -> Optional[str]:
+ return ">=1.0.0,<2.0.0"
+
+
+ class FusionBlockV1(WorkflowBlock):
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return BlockManifest
+
+ def run(
+ self,
+ predictions: List[sv.Detections],
+ ) -> BlockResult:
+ merged = sv.Detections.merge(predictions)
+ return {"predictions": merged}
+ ```
+
+ * lines `23-26` depict how to define manifest field capable of accepting
+ list of selectors
+
+ * line `50` shows what to expect as input to block's `run(...)` method -
+ list of objects which are representation of specific kind. If the block accepted
+ batches, the input type of `predictions` field would be `List[Batch[sv.Detections]`
+
+Such block is compatible with the following step declaration:
+
+```{ .json linenums="1" hl_lines="4-7"}
+{
+ "type": "my_plugin/fusion_of_predictions@v1",
+ "name": "my_step",
+ "predictions": [
+ "$steps.model_1.predictions",
+ "$steps.model_2.predictions"
+ ]
+}
+```
+
+#### Block with data transformations allowing dynamic parameters
+
+Occasionally, blocks may need to accept group of "named" selectors,
+which names and values are to be defined by creator of Workflow definition.
+In such cases, block manifest shall accept dictionary of selectors, where
+keys serve as names for those selectors.
+
+??? example "Nested selectors - named selectors"
+
+ ```{ .py linenums="1" hl_lines="23-26 47"}
+ from typing import List, Literal, Optional, Type, Any
+
+ from pydantic import Field
+ import supervision as sv
+ from inference.core.workflows.execution_engine.entities.base import (
+ OutputDefinition,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ StepOutputSelector,
+ WorkflowParameterSelector,
+ )
+ from inference.core.workflows.prototypes.block import (
+ BlockResult,
+ WorkflowBlock,
+ WorkflowBlockManifest,
+ )
+
+
+
+ class BlockManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/named_selectors_example@v1"]
+ name: str
+ data: Dict[str, StepOutputSelector(), WorkflowParameterSelector()] = Field(
+ description="Selectors to step outputs",
+ examples=[{"a": $steps.model_1.predictions", "b": "$Inputs.data"}],
+ )
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [
+ OutputDefinition(name="my_output", kind=[]),
+ ]
+
+ @classmethod
+ def get_execution_engine_compatibility(cls) -> Optional[str]:
+ return ">=1.0.0,<2.0.0"
+
+
+ class BlockWithNamedSelectorsV1(WorkflowBlock):
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return BlockManifest
+
+ def run(
+ self,
+ data: Dict[str, Any],
+ ) -> BlockResult:
+ ...
+ return {"my_output": ...}
+ ```
+
+ * lines `23-26` depict how to define manifest field capable of accepting
+ list of selectors
+
+ * line `47` shows what to expect as input to block's `run(...)` method -
+ dict of objects which are reffered with selectors. If the block accepted
+ batches, the input type of `data` field would be `Dict[str, Union[Batch[Any], Any]]`.
+ In non-batch cases, non-batch-oriented data referenced by selector is automatically
+ broadcasted, whereas for blocks accepting batches - `Batch` container wraps only
+ batch-oriented inputs, with other inputs being passed as singular values.
+
+Such block is compatible with the following step declaration:
+
+```{ .json linenums="1" hl_lines="4-7"}
+{
+ "type": "my_plugin/named_selectors_example@v1",
+ "name": "my_step",
+ "data": {
+ "a": "$steps.model_1.predictions",
+ "b": "$inputs.my_parameter"
+ }
+}
+```
+
+Practical implications will be the following:
+
+* under `data["a"]` inside `run(...)` you will be able to find model's predictions -
+like `sv.Detections` if `model_1` is object-detection model
+
+* under `data["b"]` inside `run(...)`, you will find value of input parameter named `my_parameter`
+
+### Inputs and output dimensionality vs `run(...)` method
+
+The dimensionality of block inputs plays a crucial role in shaping the `run(...)` method’s signature, and that's
+why the system enforces strict bounds on the differences in dimensionality levels between inputs
+(with the maximum allowed difference being `1`). This restriction is critical for ensuring consistency and
+predictability when writing blocks.
+
+If dimensionality differences weren't controlled, it would be difficult to predict the structure of
+the `run(...)` method, making development harder and less reliable. That’s why validation of this property
+is strictly enforced during the Workflow compilation process.
+
+Similarly, the output dimensionality also affects the method signature and the format of the expected output.
+The ecosystem supports the following scenarios:
+
+* all inputs have **the same dimensionality** and outputs **does not change** dimensionality - baseline case
+
+* all inputs have **the same dimensionality** and output **decreases** dimensionality
+
+* all inputs have **the same dimensionality** and output **increases** dimensionality
+
+* inputs have **different dimensionality** and output is allowed to keep the dimensionality of
+**reference input**
+
+Other combinations of input/output dimensionalities are not allowed to ensure consistency and to prevent ambiguity in
+the method signatures.
+
+??? example "Impact of dimensionality on `run(...)` method - batches disabled"
+
+ === "output dimensionality increase"
+
+ In this example, we perform dynamic crop of image based on predictions.
+
+ ```{ .py linenums="1" hl_lines="30-32 65 66-67"}
+ from typing import Dict, List, Literal, Optional, Type, Union
+ from uuid import uuid4
+
+ from inference.core.workflows.execution_engine.constants import DETECTION_ID_KEY
+ from inference.core.workflows.execution_engine.entities.base import (
+ OutputDefinition,
+ WorkflowImageData,
+ ImageParentMetadata,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ BATCH_OF_IMAGES_KIND,
+ BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND,
+ StepOutputImageSelector,
+ StepOutputSelector,
+ WorkflowImageSelector,
+ )
+ from inference.core.workflows.prototypes.block import (
+ BlockResult,
+ WorkflowBlock,
+ WorkflowBlockManifest,
+ )
+
+ class BlockManifest(WorkflowBlockManifest):
+ type: Literal["my_block/dynamic_crop@v1"]
+ image: Union[WorkflowImageSelector, StepOutputImageSelector]
+ predictions: StepOutputSelector(
+ kind=[BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND],
+ )
+
+ @classmethod
+ def get_output_dimensionality_offset(cls) -> int:
+ return 1
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [
+ OutputDefinition(name="crops", kind=[BATCH_OF_IMAGES_KIND]),
+ ]
+
+ @classmethod
+ def get_execution_engine_compatibility(cls) -> Optional[str]:
+ return ">=1.0.0,<2.0.0"
+
+ class DynamicCropBlockV1(WorkflowBlock):
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return BlockManifest
+
+ def run(
+ self,
+ image: WorkflowImageData,
+ predictions: sv.Detections,
+ ) -> BlockResult:
+ crops = []
+ for (x_min, y_min, x_max, y_max) in predictions.xyxy.round().astype(dtype=int):
+ cropped_image = image.numpy_image[y_min:y_max, x_min:x_max]
+ parent_metadata = ImageParentMetadata(parent_id=f"{uuid4()}")
+ if cropped_image.size:
+ result = WorkflowImageData(
+ parent_metadata=parent_metadata,
+ numpy_image=cropped_image,
+ )
+ else:
+ result = None
+ crops.append({"crops": result})
+ return crops
+ ```
+
+ * in lines `30-32` manifest class declares output dimensionality
+ offset - value `1` should be understood as adding `1` to dimensionality level
+
+ * point out, that in line `65`, block eliminates empty images from further processing but
+ placing `None` instead of dictionatry with outputs. This would utilise the same
+ Execution Engine behaviour that is used for conditional execution - datapoint will
+ be eliminated from downstream processing (unless steps requesting empty inputs
+ are present down the line).
+
+ * in lines `66-67` results for single input `image` and `predictions` are collected -
+ it is meant to be list of dictionares containing all registered outputs as keys. Execution
+ engine will understand that the step returns batch of elements for each input element and
+ create nested sturcures of indices to keep track of during execution of downstream steps.
+
+ === "output dimensionality decrease"
+
+ In this example, the block visualises crops predictions and creates tiles
+ presenting all crops predictions in single output image.
+
+ ```{ .py linenums="1" hl_lines="31-33 50-51 61-62"}
+ from typing import List, Literal, Type, Union
+
+ import supervision as sv
+
+ from inference.core.workflows.execution_engine.entities.base import (
+ Batch,
+ OutputDefinition,
+ WorkflowImageData,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ BATCH_OF_IMAGES_KIND,
+ BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND,
+ StepOutputImageSelector,
+ StepOutputSelector,
+ WorkflowImageSelector,
+ )
+ from inference.core.workflows.prototypes.block import (
+ BlockResult,
+ WorkflowBlock,
+ WorkflowBlockManifest,
+ )
+
+
+ class BlockManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/tile_detections@v1"]
+ crops: Union[WorkflowImageSelector, StepOutputImageSelector]
+ crops_predictions: StepOutputSelector(
+ kind=[BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND]
+ )
+
+ @classmethod
+ def get_output_dimensionality_offset(cls) -> int:
+ return -1
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [
+ OutputDefinition(name="visualisations", kind=[BATCH_OF_IMAGES_KIND]),
+ ]
+
+
+ class TileDetectionsBlock(WorkflowBlock):
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return BlockManifest
+
+ def run(
+ self,
+ crops: Batch[WorkflowImageData],
+ crops_predictions: Batch[sv.Detections],
+ ) -> BlockResult:
+ annotator = sv.BoundingBoxAnnotator()
+ visualisations = []
+ for image, prediction in zip(crops, crops_predictions):
+ annotated_image = annotator.annotate(
+ image.numpy_image.copy(),
+ prediction,
+ )
+ visualisations.append(annotated_image)
+ tile = sv.create_tiles(visualisations)
+ return {"visualisations": tile}
+ ```
+
+ * in lines `31-33` manifest class declares output dimensionality
+ offset - value `-1` should be understood as decreasing dimensionality level by `1`
+
+ * in lines `50-51` you can see the impact of output dimensionality decrease
+ on the method signature. Both inputs are artificially wrapped in `Batch[]` container.
+ This is done by Execution Engine automatically on output dimensionality decrease when
+ all inputs have the same dimensionality to enable access to all elements occupying
+ the last dimensionality level. Obviously, only elements related to the same element
+ from top-level batch will be grouped. For instance, if you had two input images that you
+ cropped - crops from those two different images will be grouped separately.
+
+ * lines `61-62` illustrate how output is constructed - single value is returned and that value
+ will be indexed by Execution Engine in output batch with reduced dimensionality
+
+ === "different input dimensionalities"
+
+ In this example, block merges detections which were predicted based on
+ crops of original image - result is to provide single detections with
+ all partial ones being merged.
+
+ ```{ .py linenums="1" hl_lines="32-37 39-41 63-64 70"}
+ from copy import deepcopy
+ from typing import Dict, List, Literal, Optional, Type, Union
+
+ import numpy as np
+ import supervision as sv
+
+ from inference.core.workflows.execution_engine.entities.base import (
+ Batch,
+ OutputDefinition,
+ WorkflowImageData,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND,
+ StepOutputImageSelector,
+ StepOutputSelector,
+ WorkflowImageSelector,
+ )
+ from inference.core.workflows.prototypes.block import (
+ BlockResult,
+ WorkflowBlock,
+ WorkflowBlockManifest,
+ )
+
+
+ class BlockManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/stitch@v1"]
+ image: Union[WorkflowImageSelector, StepOutputImageSelector]
+ image_predictions: StepOutputSelector(
+ kind=[BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND],
+ )
+
+ @classmethod
+ def get_input_dimensionality_offsets(cls) -> Dict[str, int]:
+ return {
+ "image": 0,
+ "image_predictions": 1,
+ }
+
+ @classmethod
+ def get_dimensionality_reference_property(cls) -> Optional[str]:
+ return "image"
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [
+ OutputDefinition(
+ name="predictions",
+ kind=[
+ BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND,
+ ],
+ ),
+ ]
+
+
+ class StitchDetectionsNonBatchBlock(WorkflowBlock):
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return BlockManifest
+
+ def run(
+ self,
+ image: WorkflowImageData,
+ image_predictions: Batch[sv.Detections],
+ ) -> BlockResult:
+ image_predictions = [deepcopy(p) for p in image_predictions if len(p)]
+ for p in image_predictions:
+ coords = p["parent_coordinates"][0]
+ p.xyxy += np.concatenate((coords, coords))
+ return {"predictions": sv.Detections.merge(image_predictions)}
+
+ ```
+
+ * in lines `32-37` manifest class declares input dimensionalities offset, indicating
+ `image` parameter being top-level and `image_predictions` being nested batch of predictions
+
+ * whenever different input dimensionalities are declared, dimensionality reference property
+ must be pointed (see lines `39-41`) - this dimensionality level would be used to calculate
+ output dimensionality - in this particular case, we specify `image`. This choice
+ has an implication in the expected format of result - in the chosen scenario we are supposed
+ to return single dictionary with all registered outputs keys. If our choice is `image_predictions`,
+ we would return list of dictionaries (of size equal to length of `image_predictions` batch). In other worlds,
+ `get_dimensionality_reference_property(...)` which dimensionality level should be associated
+ to the output.
+
+ * lines `63-64` present impact of dimensionality offsets specified in lines `32-37`. It is clearly
+ visible that `image_predictions` is a nested batch regarding `image`. Obviously, only nested predictions
+ relevant for the specific `images` are grouped in batch and provided to the method in runtime.
+
+ * as mentioned earlier, line `70` construct output being single dictionary, as we register output
+ at dimensionality level of `image` (which was also shipped as single element)
+
+
+??? example "Impact of dimensionality on `run(...)` method - batches enabled"
+
+ === "output dimensionality increase"
+
+ In this example, we perform dynamic crop of image based on predictions.
+
+ ```{ .py linenums="1" hl_lines="31-33 35-37 57-58 72 73-75"}
+ from typing import Dict, List, Literal, Optional, Type, Union
+ from uuid import uuid4
+
+ from inference.core.workflows.execution_engine.constants import DETECTION_ID_KEY
+ from inference.core.workflows.execution_engine.entities.base import (
+ OutputDefinition,
+ WorkflowImageData,
+ ImageParentMetadata,
+ Batch,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ BATCH_OF_IMAGES_KIND,
+ BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND,
+ StepOutputImageSelector,
+ StepOutputSelector,
+ WorkflowImageSelector,
+ )
+ from inference.core.workflows.prototypes.block import (
+ BlockResult,
+ WorkflowBlock,
+ WorkflowBlockManifest,
+ )
+
+ class BlockManifest(WorkflowBlockManifest):
+ type: Literal["my_block/dynamic_crop@v1"]
+ image: Union[WorkflowImageSelector, StepOutputImageSelector]
+ predictions: StepOutputSelector(
+ kind=[BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND],
+ )
+
+ @classmethod
+ def accepts_batch_input(cls) -> bool:
+ return True
+
+ @classmethod
+ def get_output_dimensionality_offset(cls) -> int:
+ return 1
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [
+ OutputDefinition(name="crops", kind=[BATCH_OF_IMAGES_KIND]),
+ ]
+
+ @classmethod
+ def get_execution_engine_compatibility(cls) -> Optional[str]:
+ return ">=1.0.0,<2.0.0"
+
+ class DynamicCropBlockV1(WorkflowBlock):
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return BlockManifest
+
+ def run(
+ self,
+ image: Batch[WorkflowImageData],
+ predictions: Batch[sv.Detections],
+ ) -> BlockResult:
+ results = []
+ for single_image, detections in zip(image, predictions):
+ crops = []
+ for (x_min, y_min, x_max, y_max) in detections.xyxy.round().astype(dtype=int):
+ cropped_image = single_image.numpy_image[y_min:y_max, x_min:x_max]
+ parent_metadata = ImageParentMetadata(parent_id=f"{uuid4()}")
+ if cropped_image.size:
+ result = WorkflowImageData(
+ parent_metadata=parent_metadata,
+ numpy_image=cropped_image,
+ )
+ else:
+ result = None
+ crops.append({"crops": result})
+ results.append(crops)
+ return results
+ ```
+
+ * in lines `31-33` manifest declares that block accepts batches of inputs
+
+ * in lines `35-37` manifest class declares output dimensionality
+ offset - value `1` should be understood as adding `1` to dimensionality level
+
+ * in lines `57-68`, signature of input parameters reflects that the `run(...)` method
+ runs against inputs of the same dimensionality and those inputs are provided in batches
+
+ * point out, that in line `72`, block eliminates empty images from further processing but
+ placing `None` instead of dictionatry with outputs. This would utilise the same
+ Execution Engine behaviour that is used for conditional execution - datapoint will
+ be eliminated from downstream processing (unless steps requesting empty inputs
+ are present down the line).
+
+ * construction of the output, presented in lines `73-75` indicates two levels of nesting.
+ First of all, block operates on batches, so it is expected to return list of outputs, one
+ output for each input batch element. Additionally, this output element for each input batch
+ element turns out to be nested batch - hence for each input iage and prediction, block
+ generates list of outputs - elements of that list are dictionaries providing values
+ for each declared output.
+
+ === "output dimensionality decrease"
+
+ In this example, the block visualises crops predictions and creates tiles
+ presenting all crops predictions in single output image.
+
+ ```{ .py linenums="1" hl_lines="31-33 35-37 54-55 68-69"}
+ from typing import List, Literal, Type, Union
+
+ import supervision as sv
+
+ from inference.core.workflows.execution_engine.entities.base import (
+ Batch,
+ OutputDefinition,
+ WorkflowImageData,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ BATCH_OF_IMAGES_KIND,
+ BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND,
+ StepOutputImageSelector,
+ StepOutputSelector,
+ WorkflowImageSelector,
+ )
+ from inference.core.workflows.prototypes.block import (
+ BlockResult,
+ WorkflowBlock,
+ WorkflowBlockManifest,
+ )
+
+
+ class BlockManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/tile_detections@v1"]
+ images_crops: Union[WorkflowImageSelector, StepOutputImageSelector]
+ crops_predictions: StepOutputSelector(
+ kind=[BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND]
+ )
+
+ @classmethod
+ def accepts_batch_input(cls) -> bool:
+ return True
+
+ @classmethod
+ def get_output_dimensionality_offset(cls) -> int:
+ return -1
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [
+ OutputDefinition(name="visualisations", kind=[BATCH_OF_IMAGES_KIND]),
+ ]
+
+
+ class TileDetectionsBlock(WorkflowBlock):
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return BlockManifest
+
+ def run(
+ self,
+ images_crops: Batch[Batch[WorkflowImageData]],
+ crops_predictions: Batch[Batch[sv.Detections]],
+ ) -> BlockResult:
+ annotator = sv.BoundingBoxAnnotator()
+ visualisations = []
+ for image_crops, crop_predictions in zip(images_crops, crops_predictions):
+ visualisations_batch_element = []
+ for image, prediction in zip(image_crops, crop_predictions):
+ annotated_image = annotator.annotate(
+ image.numpy_image.copy(),
+ prediction,
+ )
+ visualisations_batch_element.append(annotated_image)
+ tile = sv.create_tiles(visualisations_batch_element)
+ visualisations.append({"visualisations": tile})
+ return visualisations
+ ```
+
+ * lines `31-33` manifest that block is expected to take batches as input
+
+ * in lines `35-37` manifest class declares output dimensionality
+ offset - value `-1` should be understood as decreasing dimensionality level by `1`
+
+ * in lines `54-55` you can see the impact of output dimensionality decrease
+ and batch processing on the method signature. First "layer" of `Batch[]` is a side effect of the
+ fact that manifest declared that block accepts batches of inputs. The second "layer" comes
+ from output dimensionality decrease. Execution Engine wrapps up the dimension to be reduced into
+ additional `Batch[]` container porvided in inputs, such that programmer is able to collect all nested
+ batches elements that belong to specific top-level batch element.
+
+ * lines `68-69` illustrate how output is constructed - for each top-level batch element, block
+ aggregates all crops and predictions and creates a single tile. As block accepts batches of inputs,
+ this procedure end up with one tile for each top-level batch element - hence list of dictionaries
+ is expected to be returned.
+
+ === "different input dimensionalities"
+
+ In this example, block merges detections which were predicted based on
+ crops of original image - result is to provide single detections with
+ all partial ones being merged.
+
+ ```{ .py linenums="1" hl_lines="32-34 36-41 43-45 67-68 77-78"}
+ from copy import deepcopy
+ from typing import Dict, List, Literal, Optional, Type, Union
+
+ import numpy as np
+ import supervision as sv
+
+ from inference.core.workflows.execution_engine.entities.base import (
+ Batch,
+ OutputDefinition,
+ WorkflowImageData,
+ )
+ from inference.core.workflows.execution_engine.entities.types import (
+ BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND,
+ StepOutputImageSelector,
+ StepOutputSelector,
+ WorkflowImageSelector,
+ )
+ from inference.core.workflows.prototypes.block import (
+ BlockResult,
+ WorkflowBlock,
+ WorkflowBlockManifest,
+ )
+
+
+ class BlockManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/stitch@v1"]
+ images: Union[WorkflowImageSelector, StepOutputImageSelector]
+ images_predictions: StepOutputSelector(
+ kind=[BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND],
+ )
+
+ @classmethod
+ def accepts_batch_input(cls) -> bool:
+ return True
+
+ @classmethod
+ def get_input_dimensionality_offsets(cls) -> Dict[str, int]:
+ return {
+ "image": 0,
+ "image_predictions": 1,
+ }
+
+ @classmethod
+ def get_dimensionality_reference_property(cls) -> Optional[str]:
+ return "image"
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [
+ OutputDefinition(
+ name="predictions",
+ kind=[
+ BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND,
+ ],
+ ),
+ ]
+
+
+ class StitchDetectionsBatchBlock(WorkflowBlock):
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return BlockManifest
+
+ def run(
+ self,
+ images: Batch[WorkflowImageData],
+ images_predictions: Batch[Batch[sv.Detections]],
+ ) -> BlockResult:
+ result = []
+ for image, image_predictions in zip(images, images_predictions):
+ image_predictions = [deepcopy(p) for p in image_predictions if len(p)]
+ for p in image_predictions:
+ coords = p["parent_coordinates"][0]
+ p.xyxy += np.concatenate((coords, coords))
+ merged_prediction = sv.Detections.merge(image_predictions)
+ result.append({"predictions": merged_prediction})
+ return result
+ ```
+
+ * lines `32-34` manifest that block is expected to take batches as input
+
+ * in lines `36-41` manifest class declares input dimensionalities offset, indicating
+ `image` parameter being top-level and `image_predictions` being nested batch of predictions
+
+ * whenever different input dimensionalities are declared, dimensionality reference property
+ must be pointed (see lines `43-45`) - this dimensionality level would be used to calculate
+ output dimensionality - in this particular case, we specify `image`. This choice
+ has an implication in the expected format of result - in the chosen scenario we are supposed
+ to return single dictionary for each element of `image` batch. If our choice is `image_predictions`,
+ we would return list of dictionaries (of size equal to length of nested `image_predictions` batch) for each
+ input `image` batch element.
+
+ * lines `67-68` present impact of dimensionality offsets specified in lines `36-41` as well as
+ the declararion of batch processing from lines `32-34`. First "layer" of `Batch[]` container comes
+ from the latter, nested `Batch[Batch[]]` for `images_predictions` comes from the definition of input
+ dimensionality offset. It is clearly visible that `image_predictions` holds batch of predictions relevant
+ for specific elements of `image` batch.
+
+ * as mentioned earlier, lines `77-78` construct output being single dictionary for each element of `image`
+ batch
+
+
+### Block accepting empty inputs
+
+As discussed earlier, some batch elements may become "empty" during the execution of a Workflow.
+This can happen due to several factors:
+
+* **Flow-control mechanisms:** Certain branches of execution can mask specific batch elements, preventing them
+from being processed in subsequent steps.
+
+* **In data-processing blocks:** In some cases, a block may not be able to produce a meaningful output for
+a specific data point. For example, a Dynamic Crop block cannot generate a cropped image if the bounding box
+size is zero.
+
+Some blocks are designed to handle these empty inputs, such as block that can replace missing outputs with default
+values. This block can be particularly useful when constructing structured outputs in a Workflow, ensuring
+that even if some elements are empty, the output lacks missing elements making it harder to parse.
+
+??? example "Block accepting empty inputs"
+
+ ```{ .py linenums="1" hl_lines="20-22 41"}
+ from typing import Any, List, Literal, Optional, Type
+
+ from inference.core.workflows.execution_engine.entities.base import (
+ Batch,
+ OutputDefinition,
+ )
+ from inference.core.workflows.execution_engine.entities.types import StepOutputSelector
+ from inference.core.workflows.prototypes.block import (
+ BlockResult,
+ WorkflowBlock,
+ WorkflowBlockManifest,
+ )
+
+
+ class BlockManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/first_non_empty_or_default@v1"]
+ data: List[StepOutputSelector()]
+ default: Any
+
+ @classmethod
+ def accepts_empty_values(cls) -> bool:
+ return True
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [OutputDefinition(name="output")]
+
+ @classmethod
+ def get_execution_engine_compatibility(cls) -> Optional[str]:
+ return ">=1.0.0,<2.0.0"
+
+
+ class FirstNonEmptyOrDefaultBlockV1(WorkflowBlock):
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return BlockManifest
+
+ def run(
+ self,
+ data: Batch[Optional[Any]],
+ default: Any,
+ ) -> BlockResult:
+ result = default
+ for data_element in data:
+ if data_element is not None:
+ return {"output": data_element}
+ return {"output": result}
+ ```
+
+ * in lines `20-22` you may find declaration stating that block acccepts empt inputs
+
+ * a consequence of lines `20-22` is visible in line `41`, when signature states that
+ input `Batch` may contain empty elements that needs to be handled. In fact - the block
+ generates "artificial" output substituting empty value, which makes it possible for
+ those outputs to be "visible" for blocks not accepting empty inputs that refer to the
+ output of this block. You should assume that each input that is substituted by Execution
+ Engine with data generated in runtime may provide optional elements.
+
+
+### Block with custom constructor parameters
+
+Some blocks may require objects constructed by outside world to work. In such
+scenario, Workflows Execution Engine job is to transfer those entities to the block,
+making it possible to be used. The mechanism is described in
+[the page presenting Workflows Compiler](/workflows/workflows_compiler/), as this is the
+component responsible for dynamic construction of steps from blocks classes.
+
+Constructor parameters must be:
+
+* requested by block - using class method `WorkflowBlock.get_init_parameters(...)`
+
+* provided in the environment running Workflows Execution Engine:
+
+ * directly, as shown in [this](/workflows/modes_of_running/#workflows-in-python-package) example
+
+ * using defaults [registered for Workflow plugin](/workflows/blocks_bundling)
+
+Let's see how to request init parameters while defining block.
+
+??? example "Block requesting constructor parameters"
+
+ ```{ .py linenums="1" hl_lines="30-31 33-35"}
+ from typing import Any, List, Literal, Optional, Type
+
+ from inference.core.workflows.execution_engine.entities.base import (
+ Batch,
+ OutputDefinition,
+ )
+ from inference.core.workflows.execution_engine.entities.types import StepOutputSelector
+ from inference.core.workflows.prototypes.block import (
+ BlockResult,
+ WorkflowBlock,
+ WorkflowBlockManifest,
+ )
+
+
+ class BlockManifest(WorkflowBlockManifest):
+ type: Literal["my_plugin/example@v1"]
+ data: List[StepOutputSelector()]
+
+ @classmethod
+ def describe_outputs(cls) -> List[OutputDefinition]:
+ return [OutputDefinition(name="output")]
+
+ @classmethod
+ def get_execution_engine_compatibility(cls) -> Optional[str]:
+ return ">=1.0.0,<2.0.0"
+
+
+ class ExampleBlock(WorkflowBlock):
+
+ def __init__(my_parameter: int):
+ self._my_parameter = my_parameter
+
+ @classmethod
+ def get_init_parameters(cls) -> List[str]:
+ return ["my_parameter"]
+
+ @classmethod
+ def get_manifest(cls) -> Type[WorkflowBlockManifest]:
+ return BlockManifest
+
+ def run(
+ self,
+ data: Batch[Any],
+ ) -> BlockResult:
+ pass
+ ```
+
+ * lines `30-31` declare class constructor which is not parameter-free
+
+ * to inform Execution Engine that block requires custom initialisation,
+ `get_init_parameters(...)` method in lines `33-35` enlists names of all
+ parameters that must be provided
diff --git a/docs/workflows/custom_python_code_blocks.md b/docs/workflows/custom_python_code_blocks.md
new file mode 100644
index 0000000000..052d485edd
--- /dev/null
+++ b/docs/workflows/custom_python_code_blocks.md
@@ -0,0 +1,344 @@
+# Dynamic Python blocks
+
+When the syntax for Workflow definitions was [outlined](/workflows/definitions/), one key
+aspect was not covered: the ability to define blocks directly within the Workflow definition itself. This section can
+include the manifest and Python code for blocks defined in-place, which are dynamically interpreted by the
+Execution Engine. These in-place blocks function similarly to those statically defined in
+[plugins](/workflows/workflows_bundling/), yet provide much more flexibility.
+
+
+!!! Warning
+
+ Dynamic blocks only work in your local deployment of `inference` and are not supported
+ on the Roboflow hosted platform.
+
+ If you wish to disable the functionality, `export ALLOW_CUSTOM_PYTHON_EXECUTION_IN_WORKFLOWS=False`
+
+## Theory
+
+The high-level overview of Dynamic Python blocks functionality:
+
+* user provides definition of dynamic block in JSON
+
+* definition contains information required by Execution Engine to
+construct `WorkflowBlockManifest` and `WorkflowBlock` out of the
+document
+
+* in runtime, Compiler turns definition into dynamically created
+Python classes - exactly the same as statically defined blocks
+
+* In Workflow definition, you may declare steps that use dynamic blocks,
+as if dynamic blocks were standard static ones
+
+
+## Example
+
+Let's take a look and discuss example workflow with dynamic Python blocks.
+
+??? tip "Workflow with dynamic block"
+
+ ```json
+ {
+ "version": "1.0",
+ "inputs": [
+ {
+ "type": "WorkflowImage",
+ "name": "image"
+ }
+ ],
+ "dynamic_blocks_definitions": [
+ {
+ "type": "DynamicBlockDefinition",
+ "manifest": {
+ "type": "ManifestDescription",
+ "block_type": "OverlapMeasurement",
+ "inputs": {
+ "predictions": {
+ "type": "DynamicInputDefinition",
+ "selector_types": [
+ "step_output"
+ ]
+ },
+ "class_x": {
+ "type": "DynamicInputDefinition",
+ "value_types": [
+ "string"
+ ]
+ },
+ "class_y": {
+ "type": "DynamicInputDefinition",
+ "value_types": [
+ "string"
+ ]
+ }
+ },
+ "outputs": {
+ "overlap": {
+ "type": "DynamicOutputDefinition",
+ "kind": []
+ }
+ }
+ },
+ "code": {
+ "type": "PythonCode",
+ "run_function_code": "\ndef run(self, predictions: sv.Detections, class_x: str, class_y: str) -> BlockResult:\n bboxes_class_x = predictions[predictions.data[\"class_name\"] == class_x]\n bboxes_class_y = predictions[predictions.data[\"class_name\"] == class_y]\n overlap = []\n for bbox_x in bboxes_class_x:\n bbox_x_coords = bbox_x[0]\n bbox_overlaps = []\n for bbox_y in bboxes_class_y:\n if bbox_y[-1][\"detection_id\"] == bbox_x[-1][\"detection_id\"]:\n continue\n bbox_y_coords = bbox_y[0]\n x_min = max(bbox_x_coords[0], bbox_y_coords[0])\n y_min = max(bbox_x_coords[1], bbox_y_coords[1])\n x_max = min(bbox_x_coords[2], bbox_y_coords[2])\n y_max = min(bbox_x_coords[3], bbox_y_coords[3])\n # compute the area of intersection rectangle\n intersection_area = max(0, x_max - x_min + 1) * max(0, y_max - y_min + 1)\n box_x_area = (bbox_x_coords[2] - bbox_x_coords[0] + 1) * (bbox_x_coords[3] - bbox_x_coords[1] + 1)\n local_overlap = intersection_area / (box_x_area + 1e-5)\n bbox_overlaps.append(local_overlap)\n overlap.append(bbox_overlaps)\n return {\"overlap\": overlap}\n"
+ }
+ },
+ {
+ "type": "DynamicBlockDefinition",
+ "manifest": {
+ "type": "ManifestDescription",
+ "block_type": "MaximumOverlap",
+ "inputs": {
+ "overlaps": {
+ "type": "DynamicInputDefinition",
+ "selector_types": [
+ "step_output"
+ ]
+ }
+ },
+ "outputs": {
+ "max_value": {
+ "type": "DynamicOutputDefinition",
+ "kind": []
+ }
+ }
+ },
+ "code": {
+ "type": "PythonCode",
+ "run_function_code": "\ndef run(self, overlaps: List[List[float]]) -> BlockResult:\n max_value = -1\n for overlap in overlaps:\n for overlap_value in overlap:\n if not max_value:\n max_value = overlap_value\n else:\n max_value = max(max_value, overlap_value)\n return {\"max_value\": max_value}\n"
+ }
+ }
+ ],
+ "steps": [
+ {
+ "type": "RoboflowObjectDetectionModel",
+ "name": "model",
+ "image": "$inputs.image",
+ "model_id": "yolov8n-640"
+ },
+ {
+ "type": "OverlapMeasurement",
+ "name": "overlap_measurement",
+ "predictions": "$steps.model.predictions",
+ "class_x": "dog",
+ "class_y": "dog"
+ },
+ {
+ "type": "ContinueIf",
+ "name": "continue_if",
+ "condition_statement": {
+ "type": "StatementGroup",
+ "statements": [
+ {
+ "type": "BinaryStatement",
+ "left_operand": {
+ "type": "DynamicOperand",
+ "operand_name": "overlaps",
+ "operations": [
+ {
+ "type": "SequenceLength"
+ }
+ ]
+ },
+ "comparator": {
+ "type": "(Number) >="
+ },
+ "right_operand": {
+ "type": "StaticOperand",
+ "value": 1
+ }
+ }
+ ]
+ },
+ "evaluation_parameters": {
+ "overlaps": "$steps.overlap_measurement.overlap"
+ },
+ "next_steps": [
+ "$steps.maximum_overlap"
+ ]
+ },
+ {
+ "type": "MaximumOverlap",
+ "name": "maximum_overlap",
+ "overlaps": "$steps.overlap_measurement.overlap"
+ }
+ ],
+ "outputs": [
+ {
+ "type": "JsonField",
+ "name": "overlaps",
+ "selector": "$steps.overlap_measurement.overlap"
+ },
+ {
+ "type": "JsonField",
+ "name": "max_overlap",
+ "selector": "$steps.maximum_overlap.max_value"
+ }
+ ]
+ }
+ ```
+
+Let's start the analysis from `dynamic_blocks_definitions` - this is the part of
+Workflow Definition that provides a list of dynamic blocks. Each block contains two sections:
+
+* `manifest` - providing JSON representation of `BlockManifest` - refer [blocks development guide](/workflows/create_workflow_block/)
+
+* `code` - shipping Python code
+
+
+### Definition of block manifest
+
+Manifest definition contains several fields, including:
+
+* `block_type` - equivalent of `type` field in block manifest - must provide unique block identifier
+
+* `inputs` - dictionary with names and definitions of dynamic inputs
+
+* `outputs` - dictionary with names and definitions of dynamic outputs
+
+* `output_dimensionality_offset` - field specifies output dimensionality
+
+* `accepts_batch_input` - field dictates if input data in runtime is to be provided in batches
+by Execution Engine
+
+* `accepts_empty_values` - field deciding if empty inputs will be ignored while
+constructing step inputs
+
+In any doubt, refer to [blocks development guide](/workflows/create_workflow_block/), as
+the dynamic blocks replicates standard blocs capabilities.
+
+
+### Definition of dynamic input
+
+Dynamic inputs define fields of dynamically created block manifest. In other words,
+this is definition based on which `BlockManifest` class will be created in runtime.
+
+Each input may define the following properties:
+
+* `has_default_value` - flag to decide if dynamic manifest field has default
+
+* `default_value` - default value (used only if `has_default_value=True`
+
+* `is_optional` - flag to decide if dynamic manifest field is optional
+
+* `is_dimensionality_reference` - flag to decide if dynamic manifest field ship
+selector to be used in runtime as dimensionality reference
+
+* `dimensionality_offset` - dimensionality offset for configured input property
+of dynamic manifest
+
+* `selector_types` - type of selectors that may be used by property (one of
+`input_image`, `step_output_image`, `input_parameter`, `step_output`). Step may not
+hold selector, but then must provide definition of specific type.
+
+* `selector_data_kind` - dictionary with list of selector kinds specific for each selector type
+
+* `value_types` - definition of specific type that is to be placed in manifest -
+this field specifies typing of dynamically created manifest fields w.r.t Python types.
+Selection of types: `any`, `integer`, `float`, `boolean`, `dict`, `list`, `strig`
+
+### Definition of dynamic output
+
+Definitions of outputs are quite simple, hold optional list of `kinds` declared
+for given output.
+
+
+### Definition of Python code
+
+Python code is shipped in JSON document with the following fields:
+
+* `run_function_code` - code of `run(...)` method of your dynamic block
+
+* `run_function_name` - name of run function
+
+* `init_function_code` - optional code for your init function that will
+assemble step state - it is expected to return dictionary, which will be available for `run()`
+function under `self._init_results`
+
+* `init_function_name` - name of init function
+
+* `imports` - list of additional imports (you may only use libraries from your environment, no dependencies will be
+automatically installed)
+
+
+### How to create `run(...)` method?
+
+You must know the following:
+
+* `run(...)` function must be defined, as if that was class instance method - with
+the first argument being `self` and remaining arguments compatible with dynamic block manifest
+declared in definition of dynamic block
+
+* you should expect baseline symbols to be provided, including your import statements and
+the following:
+
+```python
+from typing import Any, List, Dict, Set, Optional
+import supervision as sv
+import numpy as np
+import math
+import time
+import json
+import os
+import requests
+import cv2
+import shapely
+from inference.core.workflows.execution_engine.entities.base import Batch, WorkflowImageData
+from inference.core.workflows.prototypes.block import BlockResult
+```
+
+So example function may look like the following (for clarity, we provide here
+Python code formatted nicely, but you must stringify the code to place it in definition):
+
+```python
+def run(self, predictions: sv.Detections, class_x: str, class_y: str) -> BlockResult:
+ bboxes_class_x = predictions[predictions.data["class_name"] == class_x]
+ bboxes_class_y = predictions[predictions.data["class_name"] == class_y]
+ overlap = []
+ for bbox_x in bboxes_class_x:
+ bbox_x_coords = bbox_x[0]
+ bbox_overlaps = []
+ for bbox_y in bboxes_class_y:
+ if bbox_y[-1]["detection_id"] == bbox_x[-1]["detection_id"]:
+ continue
+ bbox_y_coords = bbox_y[0]
+ x_min = max(bbox_x_coords[0], bbox_y_coords[0])
+ y_min = max(bbox_x_coords[1], bbox_y_coords[1])
+ x_max = min(bbox_x_coords[2], bbox_y_coords[2])
+ y_max = min(bbox_x_coords[3], bbox_y_coords[3])
+ # compute the area of intersection rectangle
+ intersection_area = max(0, x_max - x_min + 1) * max(0, y_max - y_min + 1)
+ box_x_area = (bbox_x_coords[2] - bbox_x_coords[0] + 1) * (bbox_x_coords[3] - bbox_x_coords[1] + 1)
+ local_overlap = intersection_area / (box_x_area + 1e-5)
+ bbox_overlaps.append(local_overlap)
+ overlap.append(bbox_overlaps)
+ return {"overlap": overlap}
+```
+
+### How to create `init(...)` method?
+
+Init function is supposed to build `self._init_results` dictionary.
+
+Example:
+
+```python
+
+def my_init() -> Dict[str, Any]:
+ return {"some": "value"}
+```
+
+### Usage of Dynamic Python block as step
+
+As shown in example Workflow definition, you may simply use the block
+as if that was normal block exposed through static plugin:
+
+```json
+{
+ "type": "OverlapMeasurement",
+ "name": "overlap_measurement",
+ "predictions": "$steps.model.predictions",
+ "class_x": "dog",
+ "class_y": "dog"
+}
+```
\ No newline at end of file
diff --git a/docs/workflows/definitions.md b/docs/workflows/definitions.md
index 853eb99bb4..edb1cc7998 100644
--- a/docs/workflows/definitions.md
+++ b/docs/workflows/definitions.md
@@ -1,149 +1,190 @@
-# How to create workflow definition?
+# Understanding Workflows Definitions syntax
+
+In Roboflow Workflows, the Workflow Definition is the internal "programming language". It provides a structured
+way to define how different blocks interact, specifying the necessary inputs, outputs, and configurations.
+By using this syntax, users can create workflows without UI.
+
+Let's start from examining the Workflow Definition created in [this tutorial](/workflows/create_and_run/) and
+analyse it step by step.
+
+??? Tip "Workflow definition"
+
+ ```json
+ {
+ "version": "1.0",
+ "inputs": [
+ {
+ "type": "InferenceImage",
+ "name": "image"
+ },
+ {
+ "type": "WorkflowParameter",
+ "name": "model",
+ "default_value": "yolov8n-640"
+ }
+ ],
+ "steps": [
+ {
+ "type": "roboflow_core/roboflow_object_detection_model@v1",
+ "name": "model",
+ "images": "$inputs.image",
+ "model_id": "$inputs.model"
+ },
+ {
+ "type": "roboflow_core/dynamic_crop@v1",
+ "name": "dynamic_crop",
+ "images": "$inputs.image",
+ "predictions": "$steps.model.predictions"
+ },
+ {
+ "type": "roboflow_core/roboflow_classification_model@v1",
+ "name": "model_1",
+ "images": "$steps.dynamic_crop.crops",
+ "model_id": "dog-breed-xpaq6/1"
+ },
+ {
+ "type": "roboflow_core/detections_classes_replacement@v1",
+ "name": "detections_classes_replacement",
+ "object_detection_predictions": "$steps.model.predictions",
+ "classification_predictions": "$steps.model_1.predictions"
+ },
+ {
+ "type": "roboflow_core/bounding_box_visualization@v1",
+ "name": "bounding_box_visualization",
+ "predictions": "$steps.detections_classes_replacement.predictions",
+ "image": "$inputs.image"
+ },
+ {
+ "type": "roboflow_core/label_visualization@v1",
+ "name": "label_visualization",
+ "predictions": "$steps.detections_classes_replacement.predictions",
+ "image": "$steps.bounding_box_visualization.image"
+ }
+ ],
+ "outputs": [
+ {
+ "type": "JsonField",
+ "name": "detections",
+ "coordinates_system": "own",
+ "selector": "$steps.detections_classes_replacement.predictions"
+ },
+ {
+ "type": "JsonField",
+ "name": "visualisation",
+ "coordinates_system": "own",
+ "selector": "$steps.label_visualization.image"
+ }
+ ]
+ }
+ ```
-## Workflow definition deep dive
+## Version marker
-Workflow definition is defined via a JSON document in the following format:
-```json
-{
- "definition": {
- "version": "1.0",
- "inputs": [],
- "steps": [],
- "outputs": []
- }
-}
-```
+Every Workflow Definition begins with the version parameter, which specifies the compatible version of the
+Workflows Execution Engine. Roboflow utilizes [Semantic Versioning](https://semver.org/) to manage these
+versions and maintains one version from each major release to ensure backward compatibility.
+This means that a workflow defined for Execution Engine version `1.0.0` will function with version `1.3.4` and other
+newer versions, but workflows created for more recent versions may not be compatible with earlier ones.
+
+List of Execution Engine versions loaded on the Roboflow Hosted platform is available
+[here](https://detect.roboflow.com/workflows/execution_engine/versions).
-In general, we have three main elements of definition:
-* `inputs` - the section where we define all parameters that can be passed in the execution time by `inference` user.
-* `steps` - the section where we define computation steps, their interconnections, connections to `inputs` and `outputs`.
-* `outputs` - the section where we define all fields that needs to be rendered in the final result
+## Inputs
-## How can we refer between elements of definition?
-To create a graph of computations, we need to define links between steps - in order to do it - we need to have a way to refer to specific elements. By convention, the following references are allowed: `${type_of_element}.{name_of_element}` and `${type_of_element}.{name_of_element}.{property}`.
+Our example workflow specifies two inputs:
+```json
+[
+ {
+ "type": "InferenceImage", "name": "image"
+ },
+ {
+ "type": "WorkflowParameter", "name": "model", "default_value": "yolov8n-640"
+ }
+]
+```
+This entry in definition creates two placeholders that can be filled with data while running workflow.
-Examples:
+The first placeholder is named `image` and is of type `InferenceImage`. This special input type is batch-oriented,
+meaning it can accept one or more images at runtime to be processed as a single batch. You can add multiple inputs
+of the type `InferenceImage`, and it is expected that the data provided to these placeholders will contain
+the same number of elements. Alternatively, you can mix inputs of sizes `N` and 1, where `N` represents the number
+of elements in the batch.
-* `$inputs.image` - reference to an input called `image`
-* `$steps.my_step.predictions` - reference to a step called `my_step` and its property `predictions`
-Additionally, defining **outputs**, it is allowed (since `v0.9.14`) to use wildcard selector
-(`${type_of_element}.{name_of_element}.*`) with intention to extract all properties of given step.
+The second placeholder is a straightforward `WorkflowParameter` called model. This type of input allows users to
+inject hyperparameters — such as model variants, confidence thresholds, and reference values — at runtime. The
+value is not expected to be a batch of elements, so when you provide a list, it will be interpreted as list of
+elements, rather than batch of elements, each to be processed individually.
-In the code, we usually call references **selectors**.
+More details about the nature of batch-oriented data processing in workflows can be found
+[here](/workflows/workflow_execution).
-## How can we define `inputs`?
-At the moment, the compiler supports two types of inputs `InferenceParameter` and `InferenceImage`.
-### `InferenceImage`
+## Steps
-This input is reserved to represent image or list of images. Definition format:
-```json
-{"type": "InferenceImage", "name": "my_image"}
-```
-When creating `InferenceImage` you do not point a specific image - you just create a placeholder that will be linked with other element of the graph. This placeholder will be substituted with actual image when you run the workflow graph and provide input parameter called `my_image` that can be `np.ndarray` or other formats that `inference` support, like:
+As mentioned [here](/workflows/understanding), steps are instances of Workflow blocks connected with inputs and outputs
+of other steps to dictate how data flows through the workflow. Let's see example step definition:
```json
{
- "type": "url",
- "value": "https://here.is/my/image.jpg"
+ "type": "roboflow_core/roboflow_object_detection_model@v1",
+ "name": "model",
+ "images": "$inputs.image",
+ "model_id": "$inputs.model"
}
```
-## `InferenceParameter`
-Similar to `InferenceImage` - `InferenceParameter` creates a placeholder for a parameters that can be used in runtime to alter execution of workflow graph.
-```json
-{"type": "InferenceParameter", "name": "confidence_threshold", "default_value": 0.5, "kind": [{"name": "my_custom_kind"}]}
-```
-`InferenceParameters` may be optionally defined with default values that will be used, if no actual parameter of given name is present in user-defined input while executing the workflow graph. Type of parameter is not explicitly defined, but will be checked in runtime, prior to execution based on types of parameters that steps using this parameters can accept.
-
-Concept of `kind` was introduced in `v0.9.21` - to make it possible to denote types of
-references at high level. See more [in contributor guide](./workflows_contribution.md)
+Two common properties for each step are `type` and `name`. Type tells which block to load and name gives the step
+unique identifier, based on which other steps may refer to output of given step.
-## How can we define `steps`?
-Compiler supports multiple type of steps (that will be described later), but let's see how to define a simple one, that would be responsible for making prediction from object-detection model:
-```json
-{
- "type": "ObjectDetectionModel",
- "name": "my_object_detection",
- "image": "$inputs.image",
- "model_id": "yolov8n-640"
-}
-```
-You can see that the step must have its type associated (that's how we link JSON document elements into code definitions) and name (unique within all steps). Another required parameters are `image` and `model_id`.
+Two remaining properties declare `selectors` (this is how we call references in Workflows) to inputs - `image` and
+`model`. While running the workflow, data passed into those placeholders will be provided for block to process.
-In case of `image` - we use reference to the input - that's how we create a link between parameter that will be provided in runtime and computational step. Steps parameters can be also provided as predefined values (like `model_id` in this case). Majority of parameters can be defined both as references to inputs (or outputs of other steps) and predefined values.
+Our documentation showcases what is the structure of each block and provides examples of how each block can be
+used as workflow step. Explore our blocks collection [here](/workflows/blocks) where you can find what are
+block data inputs, outputs and configuration properties.
-## How can we define `outputs`?
+Input data bindings of blocks (like `images` property) can be filled with selectors to batch-oriented inputs and
+step outputs. Configuration properties of blocks (like `model_id`) usually can be filled with either values
+hardcoded in workflow definition (they cannot be altered in runtime) or selectors to inputs of type `WorkflowParameter`.
+For instance, valid definition can be obtained when `model_id` is either `"$inputs.image"` or `yolov8n-640`.
-Definition of single output looks like that:
+Let's see now how step outputs are referred as inputs of another step:
```json
-{"type": "JsonField", "name": "predictions", "selector": "$steps.step_1.predictions"}
+{
+ "type": "roboflow_core/dynamic_crop@v1",
+ "name": "dynamic_crop",
+ "images": "$inputs.image",
+ "predictions": "$steps.model.predictions"
+}
```
-it defines a single output dictionary key (of name `predictions`) that will be created. `selector` field creates a link between step output and result. In this case, selector points `step_1` and its property - `predictions`.
-
-Additionally, optional parameter `coordinates_system` can be defined with one of two values (`"own", "parent"`). This parameter defaults to `parent` and describe the coordinate system of detections that should be used. This setting is only important in case of more complicated graphs (where we crop based on predicted detections and later on make another detections on each and every crop).
+In this particular case, `predictions` property defines output of step named `model`. Construction of selector is
+the following: `$steps.{step_name}.{step_output_name}`. Thanks to this reference, `model` step is connected with
+`dynamic_crop` and in runtime model predictions will be passed into dynamic crop and will be reference for image
+cropping procedure.
-## Example
-In the following example, we create a pipeline that at first makes classification first. Based on results (the top class), `step_2` decides which object detection model to use (if model predicts car, `step_3` will be executed, `step_4` will be used otherwise).
+## Outputs
-Result is build from the outputs of all models. Always one of field `step_3_predictions` and `step_4_predictions` will be empty due to conditional execution.
+This section of Workflow Definition specifies how response from workflow execution looks like. Definitions of
+each response field looks like that:
```json
{
- "definition": {
- "version": "1.0",
- "inputs": [
- {"type": "InferenceImage", "name": "image"},
- {"type": "InferenceParameter", "name": "confidence_threshold"}
- ],
- "steps": [
- {
- "type": "ClassificationModel",
- "name": "step_1",
- "image": "$inputs.image",
- "model_id": "vehicle-classification-eapcd/2",
- "confidence": 0.4
- },
- {
- "type": "Condition",
- "name": "step_2",
- "left": "$steps.step_1.top",
- "operator": "equal",
- "right": "Car",
- "step_if_true": "$steps.step_3",
- "step_if_false": "$steps.step_4"
- },
- {
- "type": "ObjectDetectionModel",
- "name": "step_3",
- "image": "$inputs.image",
- "model_id": "yolov8n-640",
- "confidence": 0.5,
- "iou_threshold": "$inputs.confidence_threshold"
- },
- {
- "type": "ObjectDetectionModel",
- "name": "step_4",
- "image": "$inputs.image",
- "model_id": "yolov8n-1280",
- "confidence": 0.5,
- "iou_threshold": 0.4
- }
- ],
- "outputs": [
- {"type": "JsonField", "name": "top_class", "selector": "$steps.step_1.top"},
- {"type": "JsonField", "name": "step_3_predictions", "selector": "$steps.step_3.predictions"},
- {"type": "JsonField", "name": "step_4_predictions", "selector": "$steps.step_4.predictions"}
- ]
- }
+ "type": "JsonField",
+ "name": "detections",
+ "selector": "$steps.detections_classes_replacement.predictions"
}
```
-## The notion of parents in `workflows`
-
-Let's imagine a scenario when we have a graph definition that requires inference from object detection model on input image. For each image that we have as an input - there will be most likely several detections. There is nothing that prevents us from doing something with those detections. For instance, we can crop original image to extract RoIs with objects that the model detected. For each crop, we may then apply yet another, specialised object detection model to detect lower resolution details. As you probably know, when `inference` makes prediction, it outputs the coordinates of detections scaled to the size of input image.
+The `selector` can reference either an input or a step output. Additionally, you can specify the `"coordinates_system"`
+property, which accepts two values: `"own"` or `"parent"`. This property is relevant for outputs that provide model
+detections and determines the coordinate system used for the detections. This becomes crucial when applying a
+secondary object detection model on image crops derived from predictions of a primary model. In such cases,
+the secondary model’s predictions are based on the coordinates of the crops, not the original input image.
+To ensure these coordinates are not translated back to the parent coordinate system, set
+`"coordinates_system": "own"` (`parent` is default option).
-But in this example, the input image is unknown when we start the process - those will be inferred by first model. To make it possible to combine predictions, we introduced `parent_id` identifier of prediction. It will be randomly generated string or name of input element that is responsible for certain prediction.
+Additionally, outputs selectors support wildcards (`$steps.step_nane.*"`) to grab all outputs of specific step.
-In our example, each detection from first model will be assigned unique identifier (`detection_id`). This identifier will be a `parent_id` for each prediction that is made based on the crop originated in detection. What is more, each output can be `coordinates_system` parameter deciding how to present the result. If `parent` coordinates mode is selected - detections made against crop will be translated to the coordinates of original image that was submitted. Thanks to that, results can be easily overlay on the input image (for instance using `supervision` library).
\ No newline at end of file
+To fully understand how output structure is created - read about
+[data processing in Workflows](/workflows/workflow_execution/).
diff --git a/docs/workflows/gallery_index.md b/docs/workflows/gallery_index.md
new file mode 100644
index 0000000000..0bd173c750
--- /dev/null
+++ b/docs/workflows/gallery_index.md
@@ -0,0 +1,17 @@
+# Workflows gallery
+
+The Workflows gallery offers example workflow definitions to help you understand what can be achieved with workflows.
+Browse through the various categories to find inspiration and ideas for building your own workflows.
+
+
\ No newline at end of file
diff --git a/docs/workflows/kinds.md b/docs/workflows/kinds.md
index 0e469e6b28..5b214904fa 100644
--- a/docs/workflows/kinds.md
+++ b/docs/workflows/kinds.md
@@ -1,12 +1,36 @@
# Workflows kinds
-In `workflows` - some values are not possible to be defined at the moment
-when `workflow` definition is created. That's why execution engine supports
-selectors - which define references to step outputs or workflow run inputs.
-To hint execution engine what is going to be provided once reference is
-resolved we need a simple type system - that's what we call `kinds`.
+In Workflows, some values cannot be defined when the Workflow Definition is created. To address this, the Execution
+Engine supports selectors, which are references to step outputs or workflow inputs. To help the Execution Engine
+understand what type of data will be provided once a reference is resolved, we use a simple type system known as
+`kinds`.
+
+`Kinds` are used to represent the semantic meaning of the underlying data. When a step outputs data of a specific
+`kind` and another step requires input of that same `kind`, the system assumes that the data will be compatible.
+This reduces the need for extensive type-compatibility checks.
+
+For example, we have different kinds to distinguish between predictions from `object detection` and
+`instance segmentation` models, even though representation of those `kinds` is
+[`sv.Detections(...)`](https://supervision.roboflow.com/latest/detection/core/). This distinction ensures that each
+block that needs a segmentation mask clearly indicates this requirement, avoiding the need to repeatedly check
+for the presence of a mask in the input.
+
+!!! Note
+
+ As for now, `kinds` are such simplistic that do not support types polymorphism - and developers
+ are asked to use unions of kinds to solve that problem. As defining extensive unions of kinds may be
+ problematic, this problem will probably be addressed in Execution Engine `v2`.
+
+!!! Warning
+
+ The list presented below contains elements with `Batch[X]` markers - those will
+ get soon deprecated and we will use only `X` markers. For now, developers are asked
+ to create their blocks using the `Batch[X]` markers, but raise the
+ [issue here](https://github.com/roboflow/inference/issues/608). This GH issue will be used
+ as a point of communication regarding deprecation process.
-## List of `workflows` kinds
+
+## Kinds declared in Roboflow plugins
* [`Batch[boolean]`](/workflows/kinds/batch_boolean): Boolean values batch
* [`dictionary`](/workflows/kinds/dictionary): Dictionary
diff --git a/docs/workflows/modes_of_running.md b/docs/workflows/modes_of_running.md
index 0a4272dfd2..18558c3bdc 100644
--- a/docs/workflows/modes_of_running.md
+++ b/docs/workflows/modes_of_running.md
@@ -1,26 +1,303 @@
-Workflows can be executed in `local` environment, or `remote` environment can be used. `local` means that model steps
-will be executed within the context of process running the code. `remote` will re-direct model steps into remote API
-using HTTP requests to send images and get predictions back.
-
-When `workflows` are used directly, in Python code - `compile_and_execute(...)` and `compile_and_execute_async(...)`
-functions accept `step_execution_mode` parameter that controls the execution mode.
-
-Additionally, `max_concurrent_steps` parameter dictates how many steps in parallel can be executed. This will
-improve efficiency of `remote` execution (up to the limits of remote API capacity) and can improve `local` execution
-if `model_manager` instance is capable of running parallel requests (only using extensions from
-`inference.enterprise.parallel`).
-
-There are environmental variables that controls `workflows` behaviour:
-
-* `DISABLE_WORKFLOW_ENDPOINTS` - disabling workflows endpoints from HTTP API
-* `WORKFLOWS_STEP_EXECUTION_MODE` - with values `local` and `remote` allowed to control how `workflows` are executed
-in `inference` HTTP container
-* `WORKFLOWS_REMOTE_API_TARGET` - with values `hosted` and `self-hosted` allowed - to point API to be used in `remote`
-execution mode
-* `LOCAL_INFERENCE_API_URL` will be used if `WORKFLOWS_REMOTE_API_TARGET=self-hosted` and
-`WORKFLOWS_STEP_EXECUTION_MODE=remote`
-* `WORKFLOWS_MAX_CONCURRENT_STEPS` - max concurrent steps to be allowed by `workflows` executor
-* `WORKFLOWS_REMOTE_EXECUTION_MAX_STEP_BATCH_SIZE` - max batch size for requests into remote API made when `remote`
-execution mode is chosen
-* `WORKFLOWS_REMOTE_EXECUTION_MAX_STEP_CONCURRENT_REQUESTS` - max concurrent requests to be possible in scope of
-single step execution when `remote` execution mode is chosen
+# What are the options for running workflows?
+
+There are few ways on how to run Workflow, including:
+
+- Request to HTTP API (Roboflow Hosted API or self-hosted `inference` server) running Workflows Execution Engine
+
+- Video processing using [InferencePipeline](/using_inference/inference_pipeline/)
+
+- `inference` Python package, where you can use Workflows Execution Engine directly in your Python app
+
+## HTTP API request
+
+This way of running Workflows is ideal for clients who:
+
+- Want to use Workflows as a stand-alone, independent part of their systems.
+
+- Maintain their main applications in languages other than Python.
+
+- Prefer to offload compute-heavy tasks to dedicated servers.
+
+
+Roboflow offers a hosted HTTP API that clients can use without needing their own infrastructure.
+Alternatively, the `inference` server (which can run Workflows) can be set up on-site if needed.
+
+Running Workflows with Roboflow Hosted API has several limitations:
+
+- Workflow runtime is limited to 20s
+
+- Response payload is limited to 6MB, which means that some blocks (especially visualization ones) if used
+in too large numbers, or with input images that are too large may result in failed request
+
+
+
+Integrating via HTTP is simple: just send a [request](https://detect.roboflow.com/docs#/default/infer_from_predefined_workflow__workspace_name__workflows__workflow_id__post)
+to the server. You can do this using a HTTP client library in your preferred programming language,
+leverage our Inference SDK in Python, or even use cURL. Explore the examples below to see how it’s done.
+
+!!! example "HTTP integration"
+
+ === "cURL"
+
+ To run your workflow created in Roboflow APP with `cURL`, use the following command:
+
+ ```bash
+ curl --location 'https://detect.roboflow.com/infer/workflows//' \
+ --header 'Content-Type: application/json' \
+ --data '{
+ "api_key": "",
+ "inputs": {
+ "image": {"type": "url", "value": "https://your-image-url"},
+ "parameter": "some-value"
+ }
+ }'
+ ```
+
+ Please note that:
+
+ - ``, ``, `` must be replaced with actual values -
+ valid for your Roboflow account
+
+ - keys of `inputs` dictionary are dictated by your Workflow, names may differ **dependent on
+ parameters you define**
+
+ - values of `inputs` dictionary are also dependent on your Workflow definition - inputs declared as
+ `WorkflowImage` have special structure - dictionary with `type` and `value` keys - using cURL your
+ options are `url` and `base64` as `type` - and value adjusted accordingly
+
+ === "Inference SDK in Python (Roboflow Hosted API)"
+
+ To run your workflow created in Roboflow APP with `InferenceClient`:
+
+ ```python
+ from inference_sdk import InferenceHTTPClient
+
+ client = InferenceHTTPClient(
+ api_url="https://detect.roboflow.com",
+ api_key="",
+ )
+
+ result = client.run_workflow(
+ workspace_name="",
+ workflow_id="",
+ images={
+ "image": "https://your-image-url"
+ },
+ parameters={
+ "parameter": "some-value"
+ }
+ )
+ ```
+
+ Please note that:
+
+ - ``, ``, `` must be replaced with actual values -
+ valid for your Roboflow account
+
+ - method parameter named `images` is supposed to be filled with dictionary that contains names and values
+ for all Workflow inputs declared as `WorkflowImage`. Names must match your Workflow definition,
+ as value you can pass either `np.array`, `PIL.Image`, URL to your image, local path to your image
+ or image in `base64` string. It is optional if Workflow does not define images as inputs.
+
+ - method parameter named `parameters` is supposed to be filled with dictionary that contains names and values
+ for all Workflow inputs of type `WorkflowParameter`. It's optional and must be filled according to Workflow
+ definition.
+
+ !!! note
+
+ Please make sure you have `inference-sdk` package installed in your environment
+
+ === "Inference SDK in Python (on-prem)"
+
+ To run your workflow created in Roboflow APP with `InferenceClient`:
+
+ ```python
+ from inference_sdk import InferenceHTTPClient
+
+ client = InferenceHTTPClient(
+ api_url="http://127.0.0.1:9001", # please modify that value according to URL of your server
+ api_key="",
+ )
+
+ result = client.run_workflow(
+ workspace_name="",
+ workflow_id="",
+ images={
+ "image": "https://your-image-url"
+ },
+ parameters={
+ "parameter": "some-value"
+ }
+ )
+ ```
+
+ Please note that:
+
+ - ``, ``, `` must be replaced with actual values -
+ valid for your Roboflow account
+
+ - method parameter named `images` is supposed to be filled with dictionary that contains names and values
+ for all Workflow inputs declared as `WorkflowImage`. Names must match your Workflow definition,
+ as value you can pass either `np.array`, `PIL.Image`, URL to your image, local path to your image
+ or image in `base64` string. It is optional if Workflow does not define images as inputs.
+
+ - method parameter named `parameters` is supposed to be filled with dictionary that contains names and values
+ for all Workflow inputs of type `WorkflowParameter`. It's optional and must be filled according to Workflow
+ definition.
+
+ !!! note
+
+ - Please make sure you have `inference-sdk` package installed in your environment.
+
+ - Easiest way to run `inference` server on-prem is to use `inference-cli` package command:
+ ```bash
+ inference server start
+ ```
+
+
+The above examples present how to run Workflow created and saved in Roboflow APP. It is also possible to
+create and run workflow that is created from scratch and may not contain API-KEY gated blocks (for instance
+your own blocks). Then you should use the
+[following endpoint](https://detect.roboflow.com/docs#/default/infer_from_workflow_workflows_run_post) or
+[Inference SDK](/inference_helpers/inference_sdk/#inference-workflows) as showcased in docs.
+
+
+## Video processing using `InferencePipeline`
+
+For use cases involving video files or streams, we recommend using [InferencePipeline](/using_inference/inference_pipeline/), which can run
+Workflows on each video frame.
+
+This option is ideal for clients who:
+
+- Need low-latency, high-throughput video processing.
+
+- Design workflows with single-frame processing times that meet real-time requirements (though complex workflows
+might not be suitable for real-time processing)
+
+
+Explore the example below to see how to combine `InferencePipeline` with Workflows.
+
+!!! example "Integration with InferencePipeline"
+
+ ```python
+ from inference import InferencePipeline
+ from inference.core.interfaces.camera.entities import VideoFrame
+
+ def my_sink(result: dict, video_frame: VideoFrame):
+ print(result) # here you can find dictionary with outputs from your workflow
+
+
+ # initialize a pipeline object
+ pipeline = InferencePipeline.init_with_workflow(
+ api_key="",
+ workspace_name="",
+ workflow_id="",
+ video_reference=0, # Path to video, device id (int, usually 0 for built in webcams), or RTSP stream url
+ on_prediction=my_sink,
+ image_input_name="image", # this parameter holds the name of Workflow input that represents
+ # image to be processed - please ADJUST it to your Workflow Definition
+ )
+ pipeline.start() #start the pipeline
+ pipeline.join() #wait for the pipeline thread to finish
+ ```
+
+ Please note that:
+
+ - ``, ``, `` must be replaced with actual values -
+ valid for your Roboflow account
+
+ - your Workflow must accept video frames under `image` parameter - when multiple video streams are
+ given for processing, all collected video frames will be submitted in batch under `image` parameter
+ for workflow run. `image` parameter must be single batch oriented input of your workflow
+
+ - additional (non-batch oriented) inputs for your workflow can be passed as parameter to `init_with_workflow(...)`
+ method see [docs](/docs/reference/inference/core/interfaces/stream/inference_pipeline/#inference.core.interfaces.stream.inference_pipeline.InferencePipeline.init_with_workflow)
+
+ !!! note
+
+ Make sure you have `inference` or `inference-gpu` package installed in your Python environment
+
+
+## Workflows in Python package
+
+Workflows Compiler and Execution Engine are bundled with [`inference`](https://pypi.org/project/inference/) package.
+Running Workflow directly may be ideal for clients who:
+
+- maintain their applications in Python
+
+- agree for resource-heavy computations directly in their app
+
+- want to avoid additional latency and errors related to sending HTTP requests
+
+- expect full control over Workflow execution
+
+In this scenario, you are supposed to provide all required initialisation values for blocks used in your Workflow, what
+makes this mode most technologically challenging, requiring you to understand handful of topics that we cover in
+developer guide.
+
+Here you can find example on how to run simple workflow in Python code.
+
+!!! example "Integration in Python"
+
+ ```python
+ from inference.core.env import WORKFLOWS_MAX_CONCURRENT_STEPS
+ from inference.core.managers.base import ModelManager
+ from inference.core.workflows.core_steps.common.entities import StepExecutionMode
+ from inference.core.env import MAX_ACTIVE_MODELS
+ from inference.core.managers.base import ModelManager
+ from inference.core.managers.decorators.fixed_size_cache import WithFixedSizeCache
+ from inference.core.registries.roboflow import RoboflowModelRegistry
+ from inference.models.utils import ROBOFLOW_MODEL_TYPES
+
+ # initialisation of Model registry to manage models load into memory
+ # (required by core blocks exposing Roboflow models)
+ model_registry = RoboflowModelRegistry(ROBOFLOW_MODEL_TYPES)
+ model_manager = ModelManager(model_registry=model_registry)
+ model_manager = WithFixedSizeCache(model_manager, max_size=MAX_ACTIVE_MODELS)
+
+ # workflow definition
+ OBJECT_DETECTION_WORKFLOW = {
+ "version": "1.0",
+ "inputs": [
+ {"type": "WorkflowImage", "name": "image"},
+ {"type": "WorkflowParameter", "name": "model_id"},
+ {"type": "WorkflowParameter", "name": "confidence", "default_value": 0.3},
+ ],
+ "steps": [
+ {
+ "type": "RoboflowObjectDetectionModel",
+ "name": "detection",
+ "image": "$inputs.image",
+ "model_id": "$inputs.model_id",
+ "confidence": "$inputs.confidence",
+ }
+ ],
+ "outputs": [
+ {"type": "JsonField", "name": "result", "selector": "$steps.detection.*"}
+ ],
+ }
+
+ # example init paramaters for blocks - dependent on set of blocks
+ # used in your workflow
+ workflow_init_parameters = {
+ "workflows_core.model_manager": model_manager,
+ "workflows_core.api_key": ",
+ "workflows_core.step_execution_mode": StepExecutionMode.LOCAL,
+ }
+
+ # instance of Execution Engine - init(...) method invocation triggers
+ # the compilation process
+ execution_engine = ExecutionEngine.init(
+ workflow_definition=OBJECT_DETECTION_WORKFLOW,
+ init_parameters=workflow_init_parameters,
+ max_concurrent_steps=WORKFLOWS_MAX_CONCURRENT_STEPS,
+ )
+
+ # runing the workflow
+ result = execution_engine.run(
+ runtime_parameters={
+ "image": [],
+ "model_id": "yolov8n-640",
+ }
+ )
+ ```
diff --git a/docs/workflows/testing.md b/docs/workflows/testing.md
new file mode 100644
index 0000000000..e51a6d8675
--- /dev/null
+++ b/docs/workflows/testing.md
@@ -0,0 +1,32 @@
+# Testing in Workflows
+
+Testing can be challenging when not done properly, which is why we recommend a practical approach
+for testing blocks that you create. Since a block is not a standalone element in the ecosystem, testing
+might seem complex, but with the right methodology, it becomes manageable.
+
+We suggest the following approach when adding a new block:
+
+* **Unit tests** should cover:
+
+ * Parsing of the manifest, especially when aliases are in use.
+
+ * Utility functions within the block module. If written correctly, these functions should simply
+ transform input data into output data, making them easy to test.
+
+ * The `run(...)` method should be tested unit-wise only if assembling the test is straightforward.
+ Otherwise, we recommend focusing on integration tests for Workflow definitions that include the block.
+
+ * Examples can be found [here](https://github.com/roboflow/inference/tree/main/tests/workflows/unit_tests/core_steps)
+
+* **Integration tests** should contain:
+
+ * practical use cases where the block is used in collaboration with others
+
+ * assertions for results, particularly for **model predictions**. These assertions should be based on empirical
+ verification, such as by visualizing and inspecting predictions to ensure they are accurate.
+
+ * When adopting models or inference techniques from external sources (e.g., open-source models),
+ assertions should confirm that the results are consistent with what you would get outside the Workflows ecosystem,
+ ensuring compatibility and correctness.
+
+ * Examples can be found [here](https://github.com/roboflow/inference/tree/main/tests/workflows/integration_tests/execution)
diff --git a/docs/workflows/understanding.md b/docs/workflows/understanding.md
index 7671da2a1a..f467e4e463 100644
--- a/docs/workflows/understanding.md
+++ b/docs/workflows/understanding.md
@@ -1,187 +1,96 @@
-# Roboflow `workflows`
-
-Roboflow `workflows` let you integrate your Machine Learning model into your environment without
-writing a single line of code. You only need to select which steps you need to complete desired
-actions and connect them together to create a `workflow`.
-
-
-## What makes a `workflow`?
-
-As mentioned above, `workflows` are made of `steps`. `Steps` perform specific actions (like
-making prediction from a model or registering results somewhere). They are instances of `blocks`,
-which ship the `code` (implementing what's happening behind the scene) and `manifest` (describing
-the way on how `block` can be used). Great news is you do not even need to care about the `code`.
-(If you do, that's great - `workflows` only may benefit from your skills). But the clue is -
-there are two (non mutually exclusive) archetypes of people using `workflows` - the ones
-employing existing `blocks` to work by combining them together and running as `workflow` using
-`execution engine`. The other use their programming skills to create own `block` and
-seamlessly plug them into `workflows` ecosystem such that other people can use them to create
-their `workflows`. We will cover `blocks` creation topics later on, so far let's focus on
-how you can use `workflows` in your project.
-
-While creating a `workflow` you can use graphical UI or JSON `workflow definition`. UI will ultimately generate
-JSON definition, but it is much easier to use. JSON definition can be created manually, but it is
-rather something that is needed to ship `workflow` to `execution engine` rather than a tool that
-people uses on the daily basis.
-
-The first step is picking set of blocks needed to achieve your goal. Then you define `inputs` describing
-either `images` or `parameters` that will become base for processing when `workflow` is run by `execution engine`.
-Those `inputs` are placeholders in `workflow` definition and will be substituted in runtime with actual values.
-
-Once you defined your `inputs` you need to connect them into appropriate `steps`. You can do it by drawing
-connection between `workflow input` and compatible `step` input. That action creates a `selector` in
-`workflow` definition that references the specific `input` and will be resolved by `execution engine` in runtime.
-
-On the similar note, `steps` are possible to be connected. Output of one step can be referenced as an input to
-another `step` given the pair is compatible (we will describe that later or).
-
-At the end of the day, you define `workflow outputs` by selecting and naming outputs of `steps` that you
-want to expose in result of `workflow run` that will be performed by `execution engine`.
-
-And - that's everything to know about `workflows` to hit the ground running.
-
-
-## What do I need to know to create blocks?
-
-In this section we provide the knowledge required to understand on how to create a custom `workflow block`.
-This is not the ultimate guide providing all technical details - instead, this section provides description
-for terminology used in more in-depth sections of documentation and lays the foundations required to
-understand workflows in-depth.
-
-What users define as JSON `workflow definition` is actually a high-level programming language to define
-`workflow`. All the symbols provided in `definition` must be understood and resolved by `execution engine`.
-Itself, `execution engine` consist of two components - `workflow compiler` and `workflows executor`.
-Compiler is responsible for taking the `workflow definition` and turning it into `execution graph` that
-is understood by `workflows executor`, which is responsible for running the `workflow` against specific
-input. Compilation may happen once and after that different input data may be fed to `workflows executor`
-to provide results.
-
-It is important to understand the relation between code that is run by `workflows executor` (defined within
-`workflow blocks`) and JSON `workflow definitions` which is parsed and transformed by `workflows compiler`.
-First of all - valid block ships the code via implementation of abstract class called `WorkflowBlock`.
-Implementation must define methods to run the computations, but also the methods to provide description
-for inputs and outputs of the block. The latter part is probably the difference that programmers would
-experience comparing writing a custom code vs creating `workflows block`.
-
-Block inputs are described by `block manifest` - pydantic entity that defines two obligatory fields (`name` and
-`type`) and as many additional properties as needed describing parametrisation of the `block`. pydantic
-manifest serves multiple roles in `workflows`. First of all - it is the source of syntax definition. Whatever
-is valid as part of JSON `workflow definition` comes from the structure of `manifests` declared for `blocks`.
-Pydantic also validates the syntax of `workflow definition` automatically and provides OpenAPI 3.x compatible
-schemas for `blocks` (making it possible to integrate with UI). Additionally, manifest tells `execution engine`
-what are the parameters that must be injected to invocation of function to run the `block` logic.
-
-Looking at specific example. The following manifest:
-```python
-class BlockManifest(WorkflowBlockManifest):
- model_config = ConfigDict(
- json_schema_extra={
- "description": "This block produces dynamic crops based on detections from detections-based model.",
- "docs": "https://inference.roboflow.com/workflows/crop",
- "block_type": "transformation",
- }
- )
- type: Literal["Crop"]
- image: Union[InferenceImageSelector, OutputStepImageSelector] = Field(
- description="The image to infer on",
- examples=["$inputs.image", "$steps.cropping.crops"],
- )
- predictions: StepOutputSelector(
- kind=[
- OBJECT_DETECTION_PREDICTION_KIND,
- INSTANCE_SEGMENTATION_PREDICTION_KIND,
- KEYPOINT_DETECTION_PREDICTION_KIND,
- ]
- ) = Field(
- description="Reference to predictions of detection-like model, that can be based of cropping "
- "(detection must define RoI - eg: bounding box)",
- examples=["$steps.my_object_detection_model.predictions"],
- validation_alias=AliasChoices("predictions", "detections"),
- )
-```
-defines manifest for `block` performing image cropping based on detections. One may point few elements:
-* native pydantic validation of fields is heavily in use - we want that, as thanks to this mechanism,
-we can automatically validate the syntax and provide all of necessary details about entities validation
-to the outside world (for instance to the UI)
-* custom types (`InferenceImageSelector`, `OutputStepImageSelector`) are in use and somewhat strange
-concept of `kind` is introduced - we will cover that later
-* `model_config` defining extra metadata added for auto-generated `block manifest` schema
-
-Having that manifest, at the level of `workflow definition` you can instantiate the cropping step using:
-```json
-{
- "type": "crop",
- "name": "my_step",
- "image": "$inputs.image",
- "predictions": "$steps.detection.predictions"
-}
-```
-and that definition will be automatically validated. Looking at the JSON document provided above,
-you can probably see two unusual entries - `"$inputs.image"` and `"$steps.detection.predictions"` -
-those are selectors which we use to reference something that is not possible to be defined statically
-while creating `workflow definition`, but will be accessible in runtime - like image and output
-of some previous step.
-
-On the similar note, `block` must declare its outputs - which is list of properties which will be
-returned after `block` is run. Those must be declared with name (unique within single `block`) and `kind`
-(which will be covered later).
-
-### How data flows through `workflow`?
-
-Knowing the details about blocks it's also good to understand how data is managed by `executor engine`.
-Running `workflow`, user provides actual inputs that are declared in `workflow definition`. Those are
-registered in special cache created for the run. Once steps are run - `execution engine` provides
-all required parameters to steps and register outputs in the cache. Those outputs may be referenced
-by other steps by `selectors` in their manifests. At the end of the run - all declared outputs are
-grabbed from the cache.
-
-There are two categories of data that we recognise in `workflows`. Parameters that are "singular" -
-like confidence thresholds or other hyperparameters. Those are declared for all the images that
-are used in processing, not at the level of single batch elements. There are also `workflows`
-data that are organised in batches - like input images or outputs from steps. The assumptions are:
-* static elements of `manifests` (fields that do not provide selectors) and input parameters of type
-`InferenceParameter` are "singular"
-* input of type `InferenceImage`, `manifests` fields declared as selectors (apart from step selectors)
-and **all step outputs** are assumed batch-major (expect list of things to be provided)
-
-
-### Selectors and `kind`
-
-Selectors are needed to make references between steps. You can compare them to pointers in such
-programming languages as C or C++. As you probably see - selectors are quite abstract - we do not
-even know if something that is selected (input - `$inputs.name`, step - `$steps.my_step` or step
-output `$steps.my_step.output`) even exist. That aspect can be automatically verified by `compiler`.
-But even if referred element exists - what is that thing? That would barely impossible to guess in
-general case. To solve this problem we introduced simple type system on top of `selectors` - `kinds`.
-
-Rules are quite simple. If you do not define `selector` kind - it is assumed to be `wildcard` -
-equivalent to `Any`. It would work, but `compiler` would have zero knowledge about data that should
-be shipped - hence error preventions mechanisms may not fully work. It is possible however to declare
-one or many `kind` that is expected for the `selector` (or step output - as those also declare `kinds`).
-Many alternatives provided are treated as union of types.
-
-How to define `kind`? Well - `kinds` represent some abstract concept with a name - that's it. There can be
-as general `kind` as "number" and as specific as "detection_with_tracking". `blocks` creators should use
-`kinds` defined in core of the `workflows` library or create own ones - that will fit their custom `blocks`.
-
-### Stateful nature of `blocks`
-Blocks are stateful to make it possible to compile a `workflow` and run it against series of images to
-support for use cases like tracking. It is ensured by the fact that each instance of `block` (`step`)
-is a class that have constructor and maintain it's state. Some classes would require init parameters -
-those must be declared in `block` definition and will be resolved and provided by `execution engine`.
-
-## What are the sources of `blocks`?
-
-`Blocks` can be defined withing `workflows` core library, but may also be shipped as custom plugin in form
-of python library. This library must define `blocks` and provide two additional elements to fulfill
-the contract:
-
-* `load_blocks()` function accessible in `__init__.py` of library that provides `List[BlockSpecification]`
-with details of blocks.
-
-* optional dict of `REGISTERED_INITIALIZERS` - with mapping from init parameter name to specific value or
-parameter-free method to assembly the value.
-
-To load `blocks` from `plugin` - install the library in your environment, and then add the name of library
-into environmental variable `WORKFLOWS_PLUGINS` - this should be comma separated list of libraries with `plugins`.
-And that's it - `plugin` should be automatically loaded.
\ No newline at end of file
+# The pillars of Workflows
+
+In the Roboflow Workflows ecosystem, various components work together seamlessly to power your applications.
+Some of these elements will be part of your daily interactions, while others operate behind the scenes to ensure
+smooth and efficient application performance. The core pillars of the ecosystem include:
+
+- **Workflows UI:** The intuitive interface where you design and manage your workflows.
+
+- **Workflow Definition:** An interchangeable format that serves as a program written in the "workflows" language,
+defining how your workflows operate.
+
+- **Workflows Blocks:** Modular components that perform specific tasks within your workflows, organised in plugins
+which can be easily created and injected into ecosystem.
+
+- **Workflows Compiler and Execution Engine:** The systems that compile your workflow definitions and execute them,
+ensuring everything runs smoothly in your environment of choice.
+
+
+We will explore each of these components, providing you with a foundational understanding to help you navigate and
+utilize the full potential of Roboflow Workflows effectively.
+
+## Workflows UI
+
+Traditionally, building machine learning applications involves complex coding and deep technical expertise.
+Roboflow Workflows simplifies this process in two key ways: providing pre-built blocks (which will be described later),
+and delivering user-friendly GUI.
+
+The interface allows you to design applications without needing to write code. You can easily connect components
+together and achieve your goals without a deep understanding of Python or the underlying workflow language.
+
+Thanks to UI, creating powerful machine learning solutions is straightforward and accessible, allowing you to focus
+on innovation rather than intricate programming.
+
+While not essential, the UI is a highly valuable component of the Roboflow Workflows ecosystem. At the end of the
+workflow creation process it creates workflow definition required for Compiler and Execution engine to run the workflow.
+
+
+
+
+
+
+
+## Workflow definition
+
+A workflow definition is essentially a document written in the internal programming language of Roboflow Workflows.
+It allows you to separate the design of your workflow from its execution. You can create a workflow definition once
+and run it in various environments using the Workflows Compiler and Execution Engine.
+
+You have two options for creating a workflow definition: UI to design it visually or write it from scratch
+if you’re comfortable with the workflows language. More details on writing definitions manually
+are available [here](/workflows/definitions/). For now, it's important to grasp the role of the definition
+within the ecosystem.
+
+A workflow definition is in fact JSON document which outlines:
+
+- **Inputs:** These are either images or configuration parameters that influence how the workflow operates.
+Instead of hardcoding values, inputs are placeholders that will be replaced with actual data during execution.
+
+- **Steps:** These are instances of workflow blocks. Each step takes inputs from either the workflow inputs or the
+outputs of previous steps. The sequence and connections between steps determine the execution order.
+
+- **Outputs:** specify field names in execution result and reference step outputs. During runtime, referred values
+are dynamically provide based on results of workflow execution.
+
+
+## Workflow blocks
+For users of Roboflow Workflows, blocks are essentially black boxes engineered to perform specific operations.
+They act as templates for the steps executed within a workflow, each defining its own set of inputs,
+configuration properties, and outputs.
+
+When adding a block to your workflow, you need to provide its inputs by referencing either the workflow’s input
+or the output of another step. You also specify the values for any required parameters. Once the step is incorporated,
+its outputs can be referenced by subsequent steps, allowing for seamless integration and chaining of operations.
+
+The creation of blocks is a more advanced topic, which you can explore [here](/workflows/create_workflow_block).
+It’s essential to understand that blocks are grouped in workflow plugins, which are standard Python libraries.
+Roboflow offers its own set of plugins, and community members are encouraged to create their own.
+The process of importing a plugin into your environment is detailed [here](/workflows/blocks_bundling).
+
+Feel encouraged to explore [Workflows blocks prepared by Roboflow](/workflows/blocks/).
+
+## Workflows Compiler and Execution Engine
+
+The Compiler and Execution Engine are essential components of the Roboflow Workflows ecosystem, doing the heavy
+lifting so you don't have to.
+
+Much like a traditional programming compiler or interpreter, these components translate your workflow definition —
+a program you create using reusable blocks — into a format that can be executed by a computer. The workflow definition
+acts as a blueprint, with blocks functioning like functions in programming, connected to produce the desired outcomes.
+
+Roboflow provides these tools as part of their [Inference Server](/#inference-server) (which can be deployed locally or
+accessed via the Roboflow Hosted platform), [video processing component](/using_inference/inference_pipeline/),
+and [Python package](https://pypi.org/project/inference/), making it easy to run your workflows in
+various environments.
+
+For a deeper dive into the Compiler and Execution Engine, please refer to our detailed documentation.
\ No newline at end of file
diff --git a/docs/workflows/versioning.md b/docs/workflows/versioning.md
new file mode 100644
index 0000000000..4c5bf9a460
--- /dev/null
+++ b/docs/workflows/versioning.md
@@ -0,0 +1,54 @@
+# Workflows versioning
+
+Understanding the life-cycle of Workflows ecosystem is an important topic,
+especially from blocks developer perspective. Those are rules that apply:
+
+* Workflows is part of `inference` - the package itself has a release whenever
+any of its component changes and those changes are ready to be published
+
+* Workflows Execution Engine declares it's version. The game plan is the following:
+
+ * core of workflows is capable to host multiple versions of Execution Engine -
+ for instance current stable version and development version
+
+ * stable version is maintained and new features are added until there is
+ need for new version and the new version is accepted
+
+ * since new version is fully operational, previously stable version starts
+ being deprecated - there is grace period when old version will be patched
+ with bug fixes (but new features will not be added), after that it will
+ be left as is. During grace period we will call blocks creators to upgrade
+ their plugins according to requirements of new version
+
+ * core library only maintains single Execution Engine version for each major -
+ making a promise that features within major will be non-breaking and Workflow
+ created under version `1.0.0` will be fully functional under version `1.4.3` of
+ Execution Engine
+
+* to ensure stability of the ecosystem over time:
+
+ * Each Workflow Definition declares Execution Engine version it is compatible with.
+ Since the core library only maintains single version for Execution Engine,
+ `version: 1.1.0` in Workflow Definition actually request Execution Engine in version
+ `>=1.1.0,<2.0.0`
+
+ * Each block, in its manifest should provide reasonable Execution Engine compatibility -
+ for instance - if block rely on Execution Engine feature introduced in `1.3.7` it should
+ specify `>=1.3.7,<2.0.0` as compatible versions of Engine
+
+* Workflows blocks may be optionally versioned (which we recommend and apply for Roboflow plugins).
+
+ * we propose the following naming convention for blocks' type identifiers:
+ `{plugin_name}/{block_family_name}@v{X}` to ensure good utilisation of blocks identifier
+ namespace
+
+ * we suggest to only modify specific version of the block if bug-fix is needed,
+ all other changes to block should yield new version
+
+ * each version of the block is to be submitted into new module
+ (as suggested [here](/workflows/blocks_bundling/)) - even **for the price of code duplication**
+ as we think stability is more important than DRY in this particular case
+
+ * on the similar note, we suggest each block to be as independent as possible,
+ as code which is shared across blocks, may unintentionally modify other blocks
+ destroying the stability of your Workflows
diff --git a/docs/workflows/workflow_execution.md b/docs/workflows/workflow_execution.md
new file mode 100644
index 0000000000..e7ee00fed3
--- /dev/null
+++ b/docs/workflows/workflow_execution.md
@@ -0,0 +1,362 @@
+# How Workflow execution looks like?
+
+Workflow execution is a complex subject, but you don’t need to understand every detail to get started effectively.
+Grasping some basic concepts can significantly speed up your learning process with the Workflows ecosystem.
+This document provides a clear and straightforward overview, designed to help you quickly understand the
+fundamentals and build more powerful applications.
+
+For those interested in a deeper technical understanding, we invite you to explore the developer guide
+for more detailed information.
+
+## Compilation
+
+Workflow execution begins with compiling the Workflow definition. As you know, a Workflow definition is a
+JSON document that outlines inputs, steps, outputs, and connections between elements. To turn this document
+into an executable format, it must be compiled.
+
+From the Execution Engine’s perspective, this process involves creating a computation graph and checking its
+integrity and correctness. This verification step is crucial because it helps identify and alert you to errors
+early on, making it easier and faster to debug issues. For instance, if you connect incompatible blocks, use an
+invalid selector, or create a loop in your workflow, the compiler will notify you with error messages.
+
+
+Once the compilation is complete, it means your Workflow is ready to run. This confirms that:
+
+- Your Workflow is compatible with the version of the Execution Engine in your environment.
+
+- All blocks in your Workflow were successfully loaded and initialized.
+
+- The connections between blocks are valid.
+
+- The input data you provided for the Workflow has been validated.
+
+At this point, the Execution Engine can begin execution of the Workflow.
+
+
+## Data in Workflow execution
+
+When you run a Workflow, you provide input data each time. Just like a function in programming that
+can handle different input values, a Workflow can process different pieces of data each time you run it.
+Let's see what happens with the data once you trigger Workflow execution.
+
+You provide input data substituting inputs' placeholders defined in the Workflow. These placeholders are
+referenced by steps of your Workflow using selectors. When a step runs, the actual piece of data you
+provided at that moment is used to make the computation. Its outputs can be later used by other steps, based
+on steps outputs selectors declared in Workflow definition, continuing this process until the Workflow
+completes and all outputs are generated.
+
+Apart from parameters with fixed values in the Workflow definition, the definition itself does not include
+actual data values. It simply tells the Execution Engine how to direct and handle the data you provide as input.
+
+
+## What is the data?
+
+Input data in a Workflow can be divided into two types:
+
+- Data to be processed: This can be submitted as a batch of data points.
+
+- Parameters: These are single values used for specific settings or configurations.
+
+To clarify the difference, consider this simple Python function:
+
+```python
+def is_even(number: int) -> bool:
+ return number % 2 == 0
+```
+You use this function like this, providing one number at a time:
+
+```python
+is_even(number=1)
+is_even(number=2)
+is_even(number=3)
+```
+
+The situation becomes more complex with machine learning models. Unlike a simple function like `is_even(...)`,
+which processes one number at a time, ML models often handle multiple pieces of data at once. For example,
+instead of providing just one image to a classification model, you can usually submit a list of images and
+receive predictions for all of them at once.
+
+This is different from our `is_even(...)` function, which would need to be called separately
+for each number to get a list of results. The difference comes from how ML models work, especially how
+GPUs process data - applying the same operation to many pieces of data simultaneously.
+
+
+
+The `is_even(...)` function can be adapted to handle batches of data by using a loop, like this:
+
+```python
+results = []
+for number in [1, 2, 3, 4]:
+ results.append(is_even(number))
+```
+
+In Workflows, similar methods are used to handle non-batch-oriented steps facing batch input data. But what if
+step expects batch-oriented data and is given singular data point? Let's look at inference process from example
+classification model:
+
+```python
+images = [PIL.Image(...), PIL.Image(...), PIL.Image(...), PIL.Image(...)]
+model = MyClassificationModel()
+
+predictions = model.infer(images=images, confidence_threshold=0.5)
+```
+
+As you may imagine, this code has chance to run correctly, as there is substantial difference in meaning of
+`images` and `confidence_threshold` parameter. Former is batch of data to apply single operation (prediction
+from a model) and the latter is parameter influencing the processing for all elements in the batch. Virtually,
+`confidence_threshold` gets propagated (broadcast) at each element of `images` list with the same value,
+as if `confidence_threshold` was the following list: `[0.5, 0.5, 0.5, 0.5]`.
+
+As mentioned earlier, Workflow inputs can be of two types:
+
+- `WorkflowImage`: This is similar to the images parameter in our example.
+
+- `WorkflowParameters`: This works like the confidence_threshold.
+
+When you provide a single image as a `WorkflowImage` input, it is automatically expanded to form a batch.
+If your Workflow definition includes multiple `WorkflowImage` placeholders, the actual data you provide for
+execution must have the same batch size for all these inputs. The only exception is when you submit a
+single image; it will be broadcast to fit the batch size requirements of other inputs.
+
+Currently, `WorkflowImage` is the only type of batch-oriented input you can use in Workflows.
+This was introduced because the ecosystem started in the Computer Vision field, where images are a key data type.
+However, as the field evolves and expands to include multi-modal models (LMMs) and other types of data,
+you can expect additional batch-oriented data types to be introduced in the future.
+
+
+## Steps interactions with data
+
+If we asked you about the nature of step outputs in these scenarios:
+
+- **A**: The step receives non-batch-oriented parameters as input.
+
+- **B**: The step receives batch-oriented data as input.
+
+- **C**: The step receives both non-batch-oriented parameters and batch-oriented data as input.
+
+You would likely say:
+
+- In option A, the output will be non-batch.
+
+- In options B and C, the output will be a batch. In option C, the non-batch-oriented parameters will be
+broadcast to match the batch size of the data.
+
+And you’d be correct. If you understand that, you probably only have two more concepts to understand before
+you can comfortably say you understand everything needed to successfully build and run complex Workflows.
+
+
+Let’s say you want to create a Workflow with these steps:
+
+1. Detect objects in a batch of input images.
+
+2. Crop each detected object from the images.
+
+3. Classify each cropped object with a second model to add detailed labels.
+
+Here’s what happens with the data in the cropping step:
+
+1. You start with a batch of images, let’s say you have `n` images.
+
+2. The object detection model finds a different number of objects in each image.
+
+3. The cropping step then creates new images for each detected object, resulting in a new batch of images
+for each original image.
+
+So, you end up with a nested list of images, with sizes like `[(k[1], ), (k[2], ), ... (k[n])]`, where each `k[i]`
+is a batch of images with a variable size based on the number of detections. The second model (classifier)
+will process these nested batches of cropped images. There is also nothing that stops you from going deeper
+in nested batches world.
+
+Here’s where it gets tricky, but Execution Engine simplifies this complexity. It manages the nesting of
+data virtually, so blocks always receive data in a flattened, non-nested format. This makes it easier to apply
+the same block, like an object detection model or classifier, regardless of how deeply nested your data is. But
+there is a price - the notion of `dimensionality level` which dictates which steps may be connected, which not.
+
+`dimensionality level` concept refers to the level of nesting of batch. Batch oriented Workflow inputs
+have `dimensionality level 1`, crops that we described in our example have `dimensionality level 2` and so on.
+What matters from the perspective of plugging inputs to specific step is:
+
+* the difference in `dimensionality level` across step inputs
+
+* the impact of step on `dimensionality level` of output (step may decrease, keep the same or increase dimensionality)
+
+Majority of blocks are designed to work with inputs at the same dimensionality level, not changing dimensionality of
+its outputs, with some being exceptions to that rule. In our example, predictions from object-detection model
+occupy `dimensionality level 1`, while classification results are at `dimensionality level 2`, due to the fact that
+cropping step introduced new, dynamic level of dimensionality.
+
+Now, if you can find a block that accepts both object detection predictions and classification predictions, you could
+use our predictions together only if block specifies explicitly it accepts such combination of `dimensionality levels`,
+otherwise you would end up seeing compilation error. Hopefully, there is a block you could use in this context.
+
+
+
+Detections Classes Replacement block is designed to substitute bounding boxes classes labels with predictions from
+classification model performed at crops of original image with respect to bounding boxes predicted by first model.
+
+!!! Warning
+
+ We are working hard to change it, but so far the Workflow UI in Roboflow APP is not capable of displaying the
+ concept of `dimensionality level`. We know that it is suboptimal from UX perspective and very confusing but we
+ must ask for patience until this situation gets better.
+
+
+!!! Note
+
+ Workflows Compiler keeps track of `data lineage` in Workflow definition, making it impossible to mix
+ together data at higher `dimensionality levels` that do not come from the same origin. This concept is
+ described in details in developer guide. From the user perspective it is important to understand that if
+ image is cropped based on predictions from different models (or even the same model, using cropping step twice),
+ cropping outputs despite being at the same dimensionality level cannot be used as inputs to the same step.
+
+
+## Conditional execution
+
+Let’s be honest—programmers love branching, and for good reason. It’s a common and useful construct in
+programming languages.
+
+For example, it’s easy to understand what’s happening in this code:
+
+```python
+def is_string_lower_cased(my_string: str) -> str:
+ if my_string.lower() == my_string:
+ return "String was lower-cased"
+ return "String was not lower-cased"
+```
+
+
+But what about this code?
+
+```python
+def is_string_lower_cased_batched(my_string: Batch[str]) -> str:
+ pass
+```
+
+In this case, it’s not immediately clear how branching would work with a batch of strings.
+The concept of handling decisions for a single item is straightforward, but when working with batches,
+the logic needs to account for multiple inputs at once. The problem arises due to the fact that independent
+decision must be made for each element of batch - which may lead to different execution branches for
+different elements of a batch. In such simplistic example as provided it can be easily addressed:
+
+```python
+def is_string_lower_cased_batched(my_string: Batch[str]) -> Batch[str]:
+ result = []
+ for element in my_string:
+ if element.lower() == my_string:
+ result.append("String was lower-cased")
+ else:
+ result.append("String was not lower-cased")
+ return result
+```
+
+In Workflows, however we want blocks to decide where execution goes, not implement conditional statements
+inside block body and return merged results. This is why whole mechanism of conditional execution
+emerged in Workflows Execution engine. This concept is important and has its own technical depth, but from
+user perspective there are few things important to understand:
+
+- some Workflows blocks can impact execution flow - steps made out of those blocks will be specified a bunch
+of step selectors, dictating possible next steps to be decided for **each element of batch** (non-batch oriented
+steps work as traditional if-else statements in programming)
+
+- once data element is discarded from batch by conditional execution, it will be hidden from all
+affected steps down the processing path and denoted in outputs as `None`
+
+- multiple flow-control steps may affect single next step, union of conditional execution masks will be created
+and dynamically applied
+
+- step may be not executed if there is no inputs to the step left after conditional execution logic evaluation
+
+- there are special blocks capable of merging alternative execution branches, such that data from that branches
+can be referred by single selector (for instance to build outputs). Example of such block is
+`First Non Empty Or Default` - which collapses execution branches taking first value encountered or defaulting to
+specified value if no value spotted
+
+- conditional execution usually impacts Workflow outputs - all values that are affected by branching are in
+fact optional (if special blocks filling empty values are not used) and nested results may not be filled with data,
+leaving empty (potentially nested) lists in results - see details
+in [section describing output construction](#output-construction).
+
+## Output construction
+
+The most important thing to understand is that a Workflow's output is aligned with its input regarding
+batch elements order. This means the output will always be a list of dictionaries, with each dictionary
+corresponding to an item in the input batch. This structure makes it easier to parse results and handle
+them iteratively, matching the outputs to the inputs.
+
+```python
+input_images = [...]
+workflow_results = execution_engine.run(
+ runtime_parameters={"images": input_images}
+)
+
+for image, result in zip(input_images, workflow_results):
+ pass
+```
+
+Each element of the list is a dictionary with keys specified in Workflow definition via declaration like:
+
+```json
+{"type": "JsonField", "name": "predictions", "selector": "$steps.detection.predictions"}
+```
+
+what you may expect as a value under those keys, however, is dependent on the structure of the workflow.
+All non-batch results got broadcast and placed in each and every output dictionary with the same value.
+Elements at `dimensionality level 1` will be distributed evenly, with values in each dictionary corresponding
+to the alignment of input data (predictions for input image 3, will be placed in third dictionary). Elements at
+higher `dimensionality levels` will be embedded into lists of objects of types specific to the step output
+being referred.
+
+For example, let's consider again our example with object-detection model, crops and secondary classification model.
+Assuming that predictions from object detection model are registered in the output under the name
+`"object_detection_predictions"` and results of classifier are registered as `"classifier_predictions"`, you
+may expect following output once three images are submitted as input for Workflow execution:
+
+```json
+[
+ {
+ "object_detection_predictions": "here sv.Detections object with 2 bounding boxes",
+ "classifier_predictions": [
+ {"classifier_prediction": "for first crop"},
+ {"classifier_prediction": "for second crop"}
+ ]
+ },
+ {
+ "object_detection_predictions": "empty sv.Detections",
+ "classifier_predictions": []
+ },
+ {
+ "object_detection_predictions": "here sv.Detections object with 3 bounding boxes",
+ "classifier_predictions": [
+ {"classifier_prediction": "for first crop"},
+ {"classifier_prediction": "for second crop"},
+ {"classifier_prediction": "for third crop"}
+ ]
+ }
+]
+```
+
+As you can see, `"classifier_predictions"` field is populated with list of results, of size equivalent to number
+of bounding boxes for `"object_detection_predictions"`.
+
+Interestingly, if our workflows has ContinueIf block that only runs cropping and classifier if number of bounding boxes
+is different from two - it will turn `classifier_predictions` in first dictionary into empty list. If conditional
+execution excludes steps at higher `dimensionality levels` from producing outputs as a side effect of execution -
+output field selecting that values will be presented as nested list of empty lists, with depth matching
+`dimensionality level - 1` of referred output.
+
+Some outputs would require serialisation when Workflows Execution Engine runs behind HTTP API. We use the following
+serialisation strategies:
+
+- images got serialised into `base64`
+
+- numpy arrays are serialised into lists
+
+- sv.Detections are serialised into `inference` format which can be decoded on the other end of the wire using
+`sv.Detections.from_inference(...)`
+
+!!! Note
+
+ sv.Detections, which is our standard representation of detection-based predictions is treated specially
+ by output constructor. `JsonField` output definition can specify optionally `coordinates_system` property,
+ which may enforce translation of detection coordinates into coordinates system of parent image in workflow.
+ See more in [docs page describing outputs definitions](/workflows/definitions/)
diff --git a/docs/workflows/workflows_compiler.md b/docs/workflows/workflows_compiler.md
new file mode 100644
index 0000000000..3e0fb6145c
--- /dev/null
+++ b/docs/workflows/workflows_compiler.md
@@ -0,0 +1,309 @@
+# Compilation of Workflow Definition
+
+Compilation is a process that takes a document written in a programming language, checks its correctness,
+and transforms it into a format that the execution environment can understand.
+
+A similar process happens in the Workflows ecosystem whenever you want to run a Workflow Definition.
+The Workflows Compiler performs several steps to transform a JSON document into a computation graph, which
+is then executed by the Workflows Execution Engine. While this process can be complex, understanding it can
+be helpful for developers contributing to the ecosystem. In this document, we outline key details of the
+compilation process to assist in building Workflow blocks and encourage contributions to the core Execution Engine.
+
+
+!!! Note
+
+ This document covers the design of Execution Engine `v1` (which is current stable version). Please
+ acknowledge information about [versioning](/workflows/versioning) to understand Execution Engine
+ development cycle.
+
+## Stages of compilation
+
+Workflow compilation involves several stages, including:
+
+1. Loading available blocks: Gathering all the blocks that can be used in the workflow based on
+configuration of execution environment
+
+2. Compiling dynamic blocks: Turning [dynamic blocks definitions](/workflows/custom_python_code_blocks) into
+standard Workflow Blocks
+
+3. Parsing the Workflow Definition: Reading and interpreting the JSON document that defines the workflow, detecting
+syntax errors
+
+4. Building Workflow Execution Graph: Creating a graph that defines how data will flow through the workflow
+during execution and verifying Workflow integrity
+
+5. Initializing Workflow steps from blocks: Setting up the individual workflow steps based on the available blocks,
+steps definitions and configuration of execution environment.
+
+Let's take a closer look at each of the workflow compilation steps.
+
+### Workflows blocks loading
+
+As described in the [blocks bundling guide](/workflows/blocks_bundling/), a group of Workflow blocks can be packaged
+into a workflow plugin. A plugin is essentially a standard Python library that, in its main module, exposes specific
+functions allowing Workflow Blocks to be dynamically loaded.
+
+The Workflows Compiler and Execution Engine are designed to be independent of specific Workflow Blocks, and the
+Compiler has the ability to discover and load blocks from plugins.
+
+Roboflow provides the `roboflow_core` plugin, which includes a set of basic Workflow Blocks that are always
+loaded by the Compiler, as both the Compiler and these blocks are bundled in the `inference` package.
+
+For custom plugins, once they are installed in the Python environment, they need to be referenced using an environment
+variable called `WORKFLOWS_PLUGINS`. This variable should contain the names of the Python packages that contain the
+plugins, separated by commas.
+
+For example, if you have two custom plugins, `numpy_plugin` and `pandas_plugin`, you can enable them in
+your Workflows environment by setting:
+```bash
+export WORKFLOWS_PLUGINS="numpy_plugin,pandas_plugin"
+```
+
+Both `numpy_plugin` and `pandas_plugin` **are not paths to library repositories**, but rather
+names of the main modules of libraries shipping plugins (`import numpy_plugin` must work in your
+Python environment for the plugin to be possible to be loaded).
+
+Once Compiler loads all plugins it is ready for the next stage of compilation.
+
+### Compilation of dynamic blocks
+
+!!! Note
+
+ The topic of [dynamic Python blocks](/workflows/custom_python_code_blocks) is covered
+ in separate docs page. To unerstand the content of this section you only need to know that
+ there is a way to define Workflow Blocks in-place in Workflow Definition - specifying
+ both block manifest and Python code in JSON document. This functionality only works if you
+ run Workflows Execution Engine on your hardware and is disabled ad Roboflow hosted platform.
+
+The Workflows Compiler can transform Dynamic Python Blocks, defined directly in a Workflow Definition, into
+full-fledged Workflow Blocks at runtime. The Compiler generates these block classes dynamically based on the
+block's definition, eliminating the need for developers to manually create them as they would in a plugin.
+
+Once this process is complete, the dynamic blocks are added to the pool of available Workflow Blocks. These blocks
+can then be used in the `steps` section of your Workflow Definition, just like any other standard block.
+
+### Parsing Workflow Definition
+
+Once all Workflow Blocks are loaded, the Compiler retrieves the manifest classes for each block.
+These manifests are `pydantic` data classes that define the structure of step entries in definition.
+At parsing stage, errors with Workflows Definition are alerted, for example:
+
+- usage of non-existing blocks
+
+- invalid configuration of steps
+
+- lack of required parameters for steps
+
+Thanks to `pydantic`, the Workflows Compiler doesn't need its own parser. Additionally, blocks creators use standard
+Python library to define block manifests.
+
+
+### Building Workflow Execution Graph
+
+Building the Workflow Execution graph is the most critical stage of Workflow compilation.
+Here's how it works:
+
+#### Adding Vertices
+First, each input, step and output are added as vertices in the graph, with each vertex given a special label
+for future identification. These vertices also include metadata, like marking input vertices with seeds for data
+lineage tracking (more on this later).
+
+#### Adding Edges
+After placing the vertices, the next step is to create edges between them based on the selectors defined in
+the Workflow. The Compiler examines the block manifests to determine which properties can accept selectors
+and the expected "kind" of those selectors. This enables the Compiler to detect errors in the Workflow
+definition, such as:
+
+- Providing an output kind from one step that doesn't match the expected input kind of the next step.
+
+- Referring to non-existent steps or inputs.
+
+Each edge also contains metadata indicating which input property is being fed by the output data, which is
+helpful at later stages of compilation and during execution
+
+!!! Note
+
+ Normally, step inputs "request" data from step outputs, forming an edge from Step A's output to Step B's input
+ during Step B's processing. However, [control-flow blocks](/workflows/create_workflow_block/) are an exception,
+ as they both accept data and declare other steps in the manifest, creating a special flow-control edge in the graph.
+
+#### Structural Validation
+
+Once the graph is constructed, the Compiler checks for structural issues like cycles to ensure the graph can be
+executed properly.
+
+#### Data Lineage verification
+
+Finally, data lineage properties are populated from input nodes and carried through the graph. So, what is
+data lineage? Lineage is a list of identifiers that track the creation and nesting of batches through the steps,
+determining:
+
+- the source path of data
+
+- `dimensionality level` of data
+
+- compatibility of different pieces of data that may be referred by a step - ensuring that step will only
+take corresponding batches elements from multiple sources (such that batch element index `example: (1, 2)` refers to
+the exact same piece of data when two batch-oriented inputs are connected into the step and not to some randomly
+provided batches with different lineage that does not make sense to process together)
+
+Each time a new nested batch is created by a step, a unique identifier is added to the lineage of the output.
+This allows the Compiler to track and verify if the inputs across steps are compatible.
+
+!!! Note
+
+ Fundamental assumption of data lineage is that all batch-oriented inputs are granted
+ the same lineage identifier - so implicitly it enforces all input batches to be fed with
+ data that has corresponding data-points at corresponding positions in batches. For instance,
+ if your Workflow compares `image_1` to `image_2` (and you declare those two inputs in Wofklow Definition),
+ the Compiler assumes the elements of `image_1[3]` to correspond with `image_2[3]`.
+
+
+Thanks to lineage tracking, the Compiler can detect potential mistakes. For example, if you attempt to connect two
+dynamic crop outputs to a single step's inputs, the Compiler will notice that the number of crops in each
+output may not match. This would result in nested batch elements with mismatched indices, which could lead to
+unpredictable results during execution if the situation is not prevented.
+
+!!! Tip "Example of lineage missmatch"
+
+ Imagine the following scenario:
+
+ - you declare single image input in your Workflow
+
+ - at first you perform object detection using two different models
+
+ - you use two dynamic crop steps - to crop based on first and second model predictions
+ respectivelly
+
+ - now you want to use block to compare two images features (using classical Compute Vision methods)
+
+ What would you expect to happen when you plug inputs from those two crop steps into comparison block?
+
+ - **Without** tracing the lineage you would "flatten" and "zip" those two batches and
+ pass pairs of images to comparison block - the problem is that in this case you cannot
+ determine if the comparisons between those elements actually makes sense - probably do not!
+
+ - **With** lineage tracing - Compiler knows that you attempt to feed two batches with lineages
+ that do not match regarding last nesting level and raises compilation error.
+
+ One may ask - "ok, but maybe I would like to apply secondary classifier on both crops and
+ merge results at the end to get all results in single output - is that possible?". The answer is
+ **yes** - as mentioned above, nested batches differ only at the last lineage level - so when we use
+ some blocks from "dimensionality collapse" category - we will align the results of secondary classifiers
+ into batches at `dimensionality level` 1 with matching lineage.
+
+
+As outlined in the section dedicated to [blocks development](/workflows/create_workflow_block), each block can define
+the expected dimensionality of its inputs and outputs. This refers to how the data should be structured.
+For example, if a block needs an `image` input that's one level above a batch of `predictions`, the Compiler will
+check that this requirement is met when verifying the Workflow step. If the connections between steps don’t match
+the expected dimensionality, an error will occur. Additionally, each input is also verified to ensure it is compatible
+based on data lineage. Once the step passes validation, the output dimensionality is determined and will be used to
+check compatibility with subsequent steps.
+
+It’s important to note that blocks define dimensionality requirements in relative terms, not absolute. This means
+a block specifies the difference (or offset) in dimensionality between its inputs and outputs. This approach allows
+blocks to work flexibly at any dimensionality level.
+
+
+!!! Note
+
+ In version 1, the Workflows Compiler only supports blocks that work across two different `dimensionality levels`.
+ This was done to keep the design straightforward. If there's a need for blocks that handle more
+ `dimensionality levels` in the future, we will consider expanding this support.
+
+
+#### Denoting flow-control
+
+The Workflows Compiler helps the Execution Engine manage flow-control structures in workflows. It marks specific
+attributes that allow the system to understand how flow-control impacts building inputs for certain steps and the
+execution of the workflow graph (for more details, see the
+[Execution Engine docs](/workflows/workflows_execution_engine/)).
+
+To ensure the workflow structure is correct, the Compiler checks data lineage for flow-control steps in a
+similar way as described in the section on [data-lineage verification](#data-lineage-verification).
+
+The Compiler assumes flow-control steps can affect other steps if:
+
+* **The flow-control step operates on non-batch-oriented inputs** - in this case, the flow-control step can
+either allow or prevent the connected step (and related steps) from running entirely, even if the input
+is a batch of data - all batch elements are affected.
+
+* **The flow-control step operates on batch-oriented inputs with compatible lineage** - here, the flow-control step
+can decide separately for each element in the batch which ones will proceed and which ones will be stopped.
+
+
+## Initializing Workflow steps from blocks
+
+The documentation often refers to a Workflow Step as an instance of a Workflow Block, which serves as its prototype.
+To put it simply, a Workflow Block is a class that implements specific behavior, which can be customized by
+configuration—whether it's set by the environment running the Execution Engine, the Workflow definition,
+or inputs at runtime.
+
+In programming, we create an instance of a class using a constructor, usually requiring initialization parameters.
+One the same note, Workflow Blocks are initialized by the Workflows Compiler whenever a step in the Workflow
+references that block. Some blocks may need specific initialization parameters, while others won't.
+
+When a block requires initialization parameters:
+
+* The block must declare the parameters it needs, as explained in detail in
+the [blocks development guide](/workflows/create_workflow_block)
+
+* The values for these parameters must be provided from the environment where the Workflow is being executed.
+
+* The values for these parameters must be provided from the environment where the Workflow is being executed.
+
+This second part might seem tricky, so let’s look at an example. In the [in user guide](/workflows/modes_of_running/),
+under the section showing how to integrate with Workflows using the `inference` Python package, you might come
+across code like this:
+
+```python
+[...]
+# example init paramaters for blocks - dependent on set of blocks
+# used in your workflow
+workflow_init_parameters = {
+ "workflows_core.model_manager": model_manager,
+ "workflows_core.api_key": ",
+ "workflows_core.step_execution_mode": StepExecutionMode.LOCAL,
+}
+
+# instance of Execution Engine - init(...) method invocation triggers
+# the compilation process
+execution_engine = ExecutionEngine.init(
+ ...,
+ init_parameters=workflow_init_parameters,
+ ...,
+)
+[...]
+```
+
+In this example, `workflow_init_parameters contains` values that the Compiler uses when
+initializing Workflow steps based on block requests.
+
+
+Initialization parameters (often called "init parameters") can be passed to the Compiler in two ways:
+
+* **Explicitly:** You provide specific values (numbers, strings, objects, etc.).
+
+* **Implicitly:** Default values are defined within the Workflows plugin, which can either be specific values or
+functions (taking no parameters) that generate values dynamically, such as from environmental variables.
+
+
+The dictionary `workflow_init_parameters` shows explicitly passed init parameters. The structure of the keys
+is important: `{plugin_name}.{init_parameter_name}`. You can also specify just `{init_parameter_name}`, but this
+changes how parameters are resolved.
+
+### How Parameters Are Resolved?
+
+When the Compiler looks for a block’s required init parameter, it follows this process:
+
+1. **Exact Match:** It first checks the explicitly provided parameters for an exact match to
+`{plugin_name}.{init_parameter_name}`.
+
+2. **Default Parameters:** If no match is found, it checks the plugin’s default parameters.
+
+3. **General Match:** Finally, it looks for a general match with just `{init_parameter_name}` in the explicitly
+provided parameters.
+
+This mechanism allows flexibility, as some block parameters can have default values while others must be
+provided explicitly. Additionally, it lets certain parameters be shared across different plugins.
diff --git a/docs/workflows/workflows_contribution.md b/docs/workflows/workflows_contribution.md
deleted file mode 100644
index a4dafc1767..0000000000
--- a/docs/workflows/workflows_contribution.md
+++ /dev/null
@@ -1,198 +0,0 @@
-# `workflows block` creation crash course
-
-At start, we need to see what is required to be implemented (via `block` base class interface). That would
-be the following methods:
-
-```python
-class WorkflowBlock(ABC):
-
- @classmethod
- @abstractmethod
- def get_manifest(cls) -> Type[WorkflowBlockManifest]:
- pass
-
- @abstractmethod
- async def run_locally(
- self,
- *args,
- **kwargs,
- ) -> Union[List[Dict[str, Any]], Tuple[List[Dict[str, Any]], FlowControl]]:
- pass
-```
-
-Let's start from input manifest assuming we want to build cropping `block`. We would need the following as
-input:
-
-- image - in `workflows` it may come as selector either to workflow input or other step output
-
-- predictions - predictions with bounding boxes (made against the image) - that we can use to crop
-
-Implementation:
-
-```python
-from typing import Literal, Union
-
-from pydantic import AliasChoices, ConfigDict, Field
-from inference.core.workflows.execution_engine.entities.types import (
- BATCH_OF_INSTANCE_SEGMENTATION_PREDICTION_KIND,
- BATCH_OF_KEYPOINT_DETECTION_PREDICTION_KIND,
- BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND,
- WorkflowImageSelector,
- StepOutputImageSelector,
- StepOutputSelector,
-)
-from inference.core.workflows.prototypes.block import (
- WorkflowBlockManifest,
-)
-
-
-class BlockManifest(WorkflowBlockManifest):
- model_config = ConfigDict(
- json_schema_extra={
- "description": "This block produces dynamic crops based on detections from detections-based model.",
- "docs": "https://inference.roboflow.com/workflows/crop",
- "block_type": "transformation",
- }
- )
- type: Literal["Crop"]
- image: Union[WorkflowImageSelector, StepOutputImageSelector] = Field(
- description="The image to infer on",
- examples=["$inputs.image", "$steps.cropping.crops"],
- )
- predictions: StepOutputSelector(
- kind=[
- BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND,
- BATCH_OF_INSTANCE_SEGMENTATION_PREDICTION_KIND,
- BATCH_OF_KEYPOINT_DETECTION_PREDICTION_KIND,
- ]
- ) = Field(
- description="Reference to predictions of detection-like model, that can be based of cropping "
- "(detection must define RoI - eg: bounding box)",
- examples=["$steps.my_object_detection_model.predictions"],
- validation_alias=AliasChoices("predictions", "detections"),
- )
-```
-
-As an output we are going to provide cropped images, so we need to declare that:
-
-```python
-from typing import List
-
-from inference.core.workflows.prototypes.block import (
- WorkflowBlockManifest,
-)
-from inference.core.workflows.execution_engine.entities.base import OutputDefinition
-from inference.core.workflows.execution_engine.entities.types import (
- BATCH_OF_IMAGES_KIND,
- BATCH_OF_PARENT_ID_KIND,
-)
-
-
-class BlockManifest(WorkflowBlockManifest):
- # [...] input properties hidden
-
- @classmethod
- def describe_outputs(cls) -> List[OutputDefinition]:
- return [
- OutputDefinition(name="crops", kind=[BATCH_OF_IMAGES_KIND]),
- OutputDefinition(name="parent_id", kind=[BATCH_OF_PARENT_ID_KIND]),
- ]
-```
-In the current version, it is required to define `parent_id` for each element that we output from steps.
-
-Then we define implementation starting from class method that will provide manifest:
-
-```python
-from typing import Type
-
-from inference.core.workflows.prototypes.block import (
- WorkflowBlock,
- WorkflowBlockManifest,
-)
-
-
-class DynamicCropBlock(WorkflowBlock):
-
- @classmethod
- def get_manifest(cls) -> Type[WorkflowBlockManifest]:
- return BlockManifest
-```
-
-Finally, we need to provide implementation for the logic:
-
-```python
-from typing import List, Tuple, Any
-import itertools
-import numpy as np
-
-from inference.core.workflows.prototypes.block import (
- WorkflowBlock,
-)
-from inference.core.workflows.execution_engine.v1.entities import FlowControl
-
-
-class DynamicCropBlock(WorkflowBlock):
-
- async def run_locally(
- self,
- image: List[dict],
- predictions: List[List[dict]],
- ) -> Tuple[List[Any], FlowControl]:
- decoded_images = [load_image(e) for e in image]
- decoded_images = [
- i[0] if i[1] is True else i[0][:, :, ::-1] for i in decoded_images
- ]
- origin_image_shape = extract_origin_size_from_images(
- input_images=image,
- decoded_images=decoded_images,
- )
- result = list(
- itertools.chain.from_iterable(
- crop_image(image=i, predictions=d, origin_size=o)
- for i, d, o in zip(decoded_images, predictions, origin_image_shape)
- )
- )
- if len(result) == 0:
- return result, FlowControl(mode="terminate_branch")
- return result, FlowControl(mode="pass")
-
-
-def crop_image(
- image: np.ndarray,
- predictions: List[dict],
- origin_size: dict,
-) -> List[Dict[str, Union[dict, str]]]:
- crops = []
- for detection in predictions:
- x_min, y_min, x_max, y_max = detection_to_xyxy(detection=detection)
- cropped_image = image[y_min:y_max, x_min:x_max]
- crops.append(
- {
- "crops": {
- IMAGE_TYPE_KEY: ImageType.NUMPY_OBJECT.value,
- IMAGE_VALUE_KEY: cropped_image,
- PARENT_ID_KEY: detection[DETECTION_ID_KEY],
- ORIGIN_COORDINATES_KEY: {
- CENTER_X_KEY: detection["x"],
- CENTER_Y_KEY: detection["y"],
- WIDTH_KEY: detection[WIDTH_KEY],
- HEIGHT_KEY: detection[HEIGHT_KEY],
- ORIGIN_SIZE_KEY: origin_size,
- },
- },
- "parent_id": detection[DETECTION_ID_KEY],
- }
- )
- return crops
-```
-
-Point out few details:
-- image come as list of dicts - each element is standard `inference` image description ("type" and "value" provided
-so `inference` loader can be used)
-
-- results of steps are provided as **list of dicts** - each element of that list ships two keys - `crops`
-and `parent_id` - which are exactly matching outputs that we defined previously.
-
-- we use `FlowControl` here - which is totally optional, but if result is a tuple with second element being
-`FlowControl` object - step may influence execution of `wokrflow` - in this case, we decide to `terminate_branch`
-(stop computations that follows this `step`) - given that we are not able to find any crops after processing.
\ No newline at end of file
diff --git a/docs/workflows/workflows_execution_engine.md b/docs/workflows/workflows_execution_engine.md
new file mode 100644
index 0000000000..b51d758298
--- /dev/null
+++ b/docs/workflows/workflows_execution_engine.md
@@ -0,0 +1,212 @@
+# Workflows Execution Engine in details
+
+The [compilation process](/workflows/workflows_compiler) creates a Workflow Execution graph, which
+holds all the necessary details to run a Workflow definition. In this section, we'll explain the details
+of the execution process.
+
+At a high level, the process does the following:
+
+1. **Validates runtime input:** it checks that all required placeholders from the Workflow definition are filled
+with data and ensures the data types are correct.
+
+2. **Determines execution order:** it defines the order in which the steps are executed.
+
+3. **Prepares step inputs and caches outputs:** it organizes the inputs for each step and saves the outputs
+for future use.
+
+4. **Builds the final Workflow outputs:** it assembles the overall result of the Workflow.
+
+## Validation of runtime input
+
+The Workflow definition specifies the expected inputs for Workflow execution. As discussed
+[earlier](/workflows/definitions), inputs can be either batch-oriented data to be processed by steps or parameters that
+configure the step execution. This distinction is crucial to how the Workflow runs and will be explored throughout
+this page.
+
+Starting with input validation, the Execution Engine has a dedicated component that parses and prepares the input
+for use. It recognizes batch-oriented inputs from the Workflow definition and converts them into an internal
+representation (e.g., `WorkflowImage` becomes `Batch[WorkflowImageData]`). This allows block developers to easily
+work with the data. Non-batch-oriented parameters are checked for type consistency against the block manifests
+used to create steps that require those parameters. This ensures that type errors are caught early in the
+execution process.
+
+!!! note
+
+ All batch-oriented inputs must have a size of either 1 or n. When a batch contains only a single element, it is
+ automatically broadcasted across the entire batch.
+
+
+## Determining execution order
+
+The Workflow Execution Graph is a [directed acyclic graph (DAG)](https://en.wikipedia.org/wiki/Directed_acyclic_graph),
+which allows us to determine the topological order. Topological order refers to a sequence in which Workflow steps are
+executed, ensuring that each step's dependencies are met before it runs. In other words, if a step relies on the output
+of another step, the Workflow Engine ensures that the dependency step is executed first.
+
+Additionally, the topological structure allows us to identify which steps can be executed in parallel without causing
+race conditions. Parallel execution is the default mode in the Workflows Execution Engine. This means that multiple
+independent steps, such as those used in model ensembling can run simultaneously, resulting in significant
+improvements in execution speed compared to sequential processing.
+
+
+!!! warning
+
+ Due to the parallel execution mode in the Execution Engine (and to avoid unnecessary data copying when passing
+ it to each step), we strongly urge all block developers to avoid mutating any data passed to the block's `run(...)`
+ method. If modifications are necessary, always make a copy of the input object before making changes!
+
+
+## Handling step inputs and outputs
+
+Handling step inputs and outputs is a complex task for the Execution Engine. This involves:
+
+* Differentiating between SIMD (Single Instruction, Multiple Data) and non-SIMD blocks in relation to their inputs.
+
+* Preparing step inputs while considering conditional execution and the expected input dimensionality.
+
+* Managing outputs from steps that control the flow of execution.
+
+* Registering outputs from data-processing steps, ensuring they match the output dimensionality declared by the blocks.
+
+Let’s explore each of these topics in detail.
+
+
+### SIMD vs non-SIMD steps
+
+As the definition suggests, a SIMD (Single Instruction, Multiple Data) step processes batch-oriented data, where the
+same operation is applied to each data point, potentially using non-batch-oriented parameters for configuration.
+The output from such a step is expected to be a batch of elements, preserving the order of the input batch elements.
+This applies to both regular processing steps and flow-control steps (see
+[blocks development guide](/workflows/create_workflow_block/ for more on their nature), where flow-control decisions
+affect each batch element individually.
+
+In essence, the type of data fed into the step determines whether it's SIMD or non-SIMD. If a step requests any
+batch-oriented input, it will be treated as a SIMD step.
+
+Non-SIMD steps, by contrast, are expected to deliver a single result for the input data. In the case of non-SIMD
+flow-control steps, they affect all downstream steps as a whole, rather than individually for each element in a batch.
+
+
+### Preparing step inputs
+
+Each requested input element may be batch-oriented or not. Non-batch inputs are relatively easy,
+they do not require special treatment. With batch-oriented ones, there is a lot more of a hustle.
+Execution Engine maintains indices for each batch-oriented datapoints, for instance:
+
+- if there is input images batch, each element will achieve their own unique index - let's say there is
+four input images, the batch indices will be `[(0, ), (1, ), (2, ), (3, )]`.
+
+- step output being not-nested batch will also be indexed, for instance predictions from a model for each of
+image mentioned above will also be indexed `[(0, ), (1, ), (2, ), (3, )]`.
+
+- having a block that increases `dimensionality level` - let's say a Dynamic Crop based on
+predictions from object-detection model - having 2 crops for first image, 1 for second and three for fourth -
+output of such step will be indexed in the following way: `[(0, 0), (0, 1), (1, 0), (3, 0), (3, 1), (3, 2)]`.
+
+Indexing of elements is important while gathering inputs for steps execution. Thanks to them, all batch oriented
+inputs may be aligned - such that Execution Engine will always ship prediction `(3, )` with image `(3, )` and
+crops batch of crops `[(3, 0), (3, 1), (3, 2)]` when any step requests it.
+
+Each requested input element can either be batch-oriented or non-batch. Non-batch inputs are straightforward and don't
+require special handling. However, batch-oriented inputs involve more complexity. The Execution Engine tracks indices
+for each batch-oriented data point. For example:
+
+- If there's a batch of input images, each element receives its own unique index. For a batch of four images, the
+indices would be `[(0,), (1,), (2,), (3,)]`.
+
+- A step output which do not increase dimensionality will also be indexed similarly. For example, model predictions
+for each of the four images would have indices `[(0,), (1,), (2,), (3,)]`.
+
+* If a block increases the `dimensionality_level` (e.g., a dynamic crop based on predictions from an object
+detection model), the output will be indexed differently. Suppose there are 2 crops for the first image,
+1 for the second, and 3 for the fourth. The indices for this output would be
+`[(0, 0), (0, 1), (1, 0), (3, 0), (3, 1), (3, 2)]`.
+
+Indexing is crucial for aligning inputs during step execution. The Execution Engine ensures that all batch-oriented
+inputs are synchronized. For example, it will match prediction `(3,)` with image `(3,)` and the corresponding batch
+of crops `[(3, 0), (3, 1), (3, 2)]` when a step requests them.
+
+!!! Note
+
+ Keeping data lineage in order during compilation simplifies execution. The Execution Engine doesn't need to
+ verify if dynamically created nested batches come from the same source. Its job is to align indices when
+ preparing step inputs.
+
+
+#### Additional Considerations
+
+##### Input Dimensionality Offsets
+Workflow blocks define how input dimensionality is handled.
+If the Execution Engine detects a difference in input dimensionality, it will wrap the larger dimension into a batch.
+For example, if a block processes both input images and dynamically cropped images, the latter will be wrapped into a
+batch so that each top-level image is processed with its corresponding batch of crops.
+
+##### Deeply nested batches are flattened before step execution
+Given the block defines all input at the same dimensionality level, no matter how deep the nesting of input batches is,
+step input will be flattened to a single batch and indices in the outputs will be automagically maintained by
+Execution Engine.
+
+##### Conditional Execution
+Flow-control blocks manage which steps should be executed based on certain conditions. During compilation,
+steps affected by these conditions are flagged. When constructing their inputs, a mask for flow-control exclusion
+(both SIMD- and non-SIMD-oriented) is applied. Based on this mask, specific input elements will be replaced with
+`None`, representing an empty value.
+
+By default, blocks don't accept empty values, so any `None` at index `(i,)` in a batch will cause that index to be
+excluded from processing. This is how flow control is managed within the Execution Engine. Some blocks, however,
+are designed to handle empty inputs. In such cases, while the flow-control mask will be applied, empty inputs
+won't be eliminated from the input batch.
+
+
+##### Batch Processing Mode
+Blocks may either process batch inputs all at once or, by default, require the Execution Engine to loop over each
+input and repeatedly invoke the block's `run(...)` method.
+
+### Managing flow-control steps outputs
+
+The outputs of flow-control steps are unique because these steps determine which data points should be
+passed to subsequent steps which is roughly similar to outcome of this pseudocode:
+
+```python
+if condition(A):
+ step_1(A)
+ step_2(A)
+else:
+ step_3(A)
+```
+
+The Workflows Execution Engine parses the outputs from flow-control steps and creates execution branches.
+Each branch has an associated mask:
+
+* For **SIMD branches**, the mask contains a set of indices that will remain active for processing.
+
+* For **non-SIMD branches**, the mask is a simple `True` / `False` value that determines whether the entire
+branch is active.
+
+After a flow-control step executes, this mask is registered and applied to any steps affected by the decision.
+This allows the Engine to filter out specific data points from processing in the downstream branch.
+If a data point is excluded from the first step in a branch (due to the masking), that data point is automatically
+eliminated from the entire branch (as a result of exclusion of empty inputs by default).
+
+### Caching steps outputs
+
+It's not just the outcomes of flow-control steps that need to be managed carefully—data processing steps also require
+attention to ensure their results are correctly passed to other steps. The key aspect here is properly indexing
+the outputs.
+
+In simple cases where all inputs share the same `dimensionality level` and the output maintains that same
+dimensionality, the Execution Engine's main task is to preserve the order of input indices. However, when input
+dimensionalities differ, the Workflow block used to create the step determines how indexing should be handled.
+
+If the dimensionality changes during processing, the Execution Engine either uses the high-level index or creates
+nested dimensions dynamically based on the length of element lists in the output. This ensures proper alignment and
+tracking of data across steps.
+
+
+## Building Workflow outputs
+
+
+For details on how outputs are constructed, please refer to the information provided on the
+[Workflows Definitions](/workflows/definitions/) page and the
+[Output Construction](/workflows/workflow_execution/#output-construction) section of the Workflow Execution
+documentation.
diff --git a/inference/core/interfaces/stream/inference_pipeline.py b/inference/core/interfaces/stream/inference_pipeline.py
index 3ac040b520..9155df08d1 100644
--- a/inference/core/interfaces/stream/inference_pipeline.py
+++ b/inference/core/interfaces/stream/inference_pipeline.py
@@ -133,7 +133,7 @@ def init(
to reflect changes in sink function signature.
Args:
- model_id (str): Name and version of model at Roboflow platform (example: "my-model/3")
+ model_id (str): Name and version of model on the Roboflow platform (example: "my-model/3")
video_reference (Union[str, int, List[Union[str, int]]]): Reference of source or sources to be used to make
predictions against. It can be video file path, stream URL and device (like camera) id
(we handle whatever cv2 handles). It can also be a list of references (since v0.9.18) - and then
@@ -464,7 +464,7 @@ def init_with_workflow(
api_key (Optional[str]): Roboflow API key - if not passed - will be looked in env under "ROBOFLOW_API_KEY"
and "API_KEY" variables. API key, passed in some form is required.
image_input_name (str): Name of input image defined in `workflow_specification` or Workflow definition saved
- at Roboflow Platform. `InferencePipeline` will be injecting video frames to workflow through that
+ on the Roboflow Platform. `InferencePipeline` will be injecting video frames to workflow through that
parameter name.
workflows_parameters (Optional[Dict[str, Any]]): Dictionary with additional parameters that can be
defined within `workflow_specification`.
@@ -500,7 +500,7 @@ def init_with_workflow(
canceled at the end of InferencePipeline processing. By default, when video file ends or
pipeline is stopped, tasks that has not started will be cancelled.
video_metadata_input_name (str): Name of input for video metadata defined in `workflow_specification` or
- Workflow definition saved at Roboflow Platform. `InferencePipeline` will be injecting video frames
+ Workflow definition saved on the Roboflow Platform. `InferencePipeline` will be injecting video frames
metadata to workflows through that parameter name.
Other ENV variables involved in low-level configuration:
* INFERENCE_PIPELINE_PREDICTIONS_QUEUE_SIZE - size of buffer for predictions that are ready for dispatching
diff --git a/inference/core/workflows/core_steps/models/roboflow/instance_segmentation/v1.py b/inference/core/workflows/core_steps/models/roboflow/instance_segmentation/v1.py
index 2fada617fa..e04814a512 100644
--- a/inference/core/workflows/core_steps/models/roboflow/instance_segmentation/v1.py
+++ b/inference/core/workflows/core_steps/models/roboflow/instance_segmentation/v1.py
@@ -20,6 +20,7 @@
convert_inference_detections_batch_to_sv_detections,
filter_out_unwanted_classes_from_sv_detections_batch,
)
+from inference.core.workflows.execution_engine.constants import INFERENCE_ID_KEY
from inference.core.workflows.execution_engine.entities.base import (
Batch,
OutputDefinition,
@@ -164,6 +165,7 @@ def accepts_batch_input(cls) -> bool:
@classmethod
def describe_outputs(cls) -> List[OutputDefinition]:
return [
+ OutputDefinition(name=INFERENCE_ID_KEY, kind=[STRING_KIND]),
OutputDefinition(
name="predictions",
kind=[BATCH_OF_INSTANCE_SEGMENTATION_PREDICTION_KIND],
@@ -356,6 +358,7 @@ def _post_process_result(
predictions: List[dict],
class_filter: Optional[List[str]],
) -> BlockResult:
+ inference_id = predictions[0].get(INFERENCE_ID_KEY, None)
predictions = convert_inference_detections_batch_to_sv_detections(predictions)
predictions = attach_prediction_type_info_to_sv_detections_batch(
predictions=predictions,
@@ -369,4 +372,7 @@ def _post_process_result(
images=images,
predictions=predictions,
)
- return [{"predictions": prediction} for prediction in predictions]
+ return [
+ {"inference_id": inference_id, "predictions": prediction}
+ for prediction in predictions
+ ]
diff --git a/inference/core/workflows/core_steps/models/roboflow/keypoint_detection/v1.py b/inference/core/workflows/core_steps/models/roboflow/keypoint_detection/v1.py
index e40a0b6181..1bcfcb519d 100644
--- a/inference/core/workflows/core_steps/models/roboflow/keypoint_detection/v1.py
+++ b/inference/core/workflows/core_steps/models/roboflow/keypoint_detection/v1.py
@@ -21,6 +21,7 @@
convert_inference_detections_batch_to_sv_detections,
filter_out_unwanted_classes_from_sv_detections_batch,
)
+from inference.core.workflows.execution_engine.constants import INFERENCE_ID_KEY
from inference.core.workflows.execution_engine.entities.base import (
Batch,
OutputDefinition,
@@ -34,6 +35,7 @@
LIST_OF_VALUES_KIND,
ROBOFLOW_MODEL_ID_KIND,
ROBOFLOW_PROJECT_KIND,
+ STRING_KIND,
FloatZeroToOne,
ImageInputField,
RoboflowModelField,
@@ -156,6 +158,7 @@ def accepts_batch_input(cls) -> bool:
@classmethod
def describe_outputs(cls) -> List[OutputDefinition]:
return [
+ OutputDefinition(name=INFERENCE_ID_KEY, kind=[STRING_KIND]),
OutputDefinition(
name="predictions", kind=[BATCH_OF_KEYPOINT_DETECTION_PREDICTION_KIND]
),
@@ -340,6 +343,7 @@ def _post_process_result(
predictions: List[dict],
class_filter: Optional[List[str]],
) -> BlockResult:
+ inference_id = predictions[0].get(INFERENCE_ID_KEY, None)
detections = convert_inference_detections_batch_to_sv_detections(predictions)
for prediction, image_detections in zip(predictions, detections):
add_inference_keypoints_to_sv_detections(
@@ -358,4 +362,7 @@ def _post_process_result(
images=images,
predictions=detections,
)
- return [{"predictions": image_detections} for image_detections in detections]
+ return [
+ {"inference_id": inference_id, "predictions": image_detections}
+ for image_detections in detections
+ ]
diff --git a/inference/core/workflows/core_steps/models/roboflow/multi_class_classification/v1.py b/inference/core/workflows/core_steps/models/roboflow/multi_class_classification/v1.py
index 1e516a96f6..90411587f7 100644
--- a/inference/core/workflows/core_steps/models/roboflow/multi_class_classification/v1.py
+++ b/inference/core/workflows/core_steps/models/roboflow/multi_class_classification/v1.py
@@ -14,6 +14,7 @@
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.core_steps.common.utils import attach_prediction_type_info
from inference.core.workflows.execution_engine.constants import (
+ INFERENCE_ID_KEY,
PARENT_ID_KEY,
ROOT_PARENT_ID_KEY,
)
@@ -28,6 +29,7 @@
FLOAT_ZERO_TO_ONE_KIND,
ROBOFLOW_MODEL_ID_KIND,
ROBOFLOW_PROJECT_KIND,
+ STRING_KIND,
FloatZeroToOne,
ImageInputField,
RoboflowModelField,
@@ -106,6 +108,7 @@ def accepts_batch_input(cls) -> bool:
@classmethod
def describe_outputs(cls) -> List[OutputDefinition]:
return [
+ OutputDefinition(name=INFERENCE_ID_KEY, kind=[STRING_KIND]),
OutputDefinition(
name="predictions", kind=[BATCH_OF_CLASSIFICATION_PREDICTION_KIND]
),
@@ -246,6 +249,7 @@ def _post_process_result(
images: Batch[WorkflowImageData],
predictions: List[dict],
) -> BlockResult:
+ inference_id = predictions[0].get(INFERENCE_ID_KEY, None)
predictions = attach_prediction_type_info(
predictions=predictions,
prediction_type="classification",
@@ -255,4 +259,7 @@ def _post_process_result(
prediction[ROOT_PARENT_ID_KEY] = (
image.workflow_root_ancestor_metadata.parent_id
)
- return [{"predictions": prediction} for prediction in predictions]
+ return [
+ {"inference_id": inference_id, "predictions": prediction}
+ for prediction in predictions
+ ]
diff --git a/inference/core/workflows/core_steps/models/roboflow/multi_label_classification/v1.py b/inference/core/workflows/core_steps/models/roboflow/multi_label_classification/v1.py
index cf4c1021ff..f8be9ea1ba 100644
--- a/inference/core/workflows/core_steps/models/roboflow/multi_label_classification/v1.py
+++ b/inference/core/workflows/core_steps/models/roboflow/multi_label_classification/v1.py
@@ -14,6 +14,7 @@
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.core_steps.common.utils import attach_prediction_type_info
from inference.core.workflows.execution_engine.constants import (
+ INFERENCE_ID_KEY,
PARENT_ID_KEY,
ROOT_PARENT_ID_KEY,
)
@@ -28,6 +29,7 @@
FLOAT_ZERO_TO_ONE_KIND,
ROBOFLOW_MODEL_ID_KIND,
ROBOFLOW_PROJECT_KIND,
+ STRING_KIND,
FloatZeroToOne,
ImageInputField,
RoboflowModelField,
@@ -106,9 +108,10 @@ def accepts_batch_input(cls) -> bool:
@classmethod
def describe_outputs(cls) -> List[OutputDefinition]:
return [
+ OutputDefinition(name=INFERENCE_ID_KEY, kind=[STRING_KIND]),
OutputDefinition(
name="predictions", kind=[BATCH_OF_CLASSIFICATION_PREDICTION_KIND]
- )
+ ),
]
@classmethod
@@ -243,6 +246,7 @@ def _post_process_result(
images: Batch[WorkflowImageData],
predictions: List[dict],
) -> List[dict]:
+ inference_id = predictions[0].get(INFERENCE_ID_KEY, None)
predictions = attach_prediction_type_info(
predictions=predictions,
prediction_type="classification",
@@ -252,4 +256,7 @@ def _post_process_result(
prediction[ROOT_PARENT_ID_KEY] = (
image.workflow_root_ancestor_metadata.parent_id
)
- return [{"predictions": image_detections} for image_detections in predictions]
+ return [
+ {"inference_id": inference_id, "predictions": prediction}
+ for prediction in predictions
+ ]
diff --git a/inference/core/workflows/core_steps/models/roboflow/object_detection/v1.py b/inference/core/workflows/core_steps/models/roboflow/object_detection/v1.py
index e2b2744120..fa282cdd64 100644
--- a/inference/core/workflows/core_steps/models/roboflow/object_detection/v1.py
+++ b/inference/core/workflows/core_steps/models/roboflow/object_detection/v1.py
@@ -18,6 +18,7 @@
convert_inference_detections_batch_to_sv_detections,
filter_out_unwanted_classes_from_sv_detections_batch,
)
+from inference.core.workflows.execution_engine.constants import INFERENCE_ID_KEY
from inference.core.workflows.execution_engine.entities.base import (
Batch,
OutputDefinition,
@@ -31,6 +32,7 @@
LIST_OF_VALUES_KIND,
ROBOFLOW_MODEL_ID_KIND,
ROBOFLOW_PROJECT_KIND,
+ STRING_KIND,
FloatZeroToOne,
ImageInputField,
RoboflowModelField,
@@ -145,6 +147,7 @@ def accepts_batch_input(cls) -> bool:
@classmethod
def describe_outputs(cls) -> List[OutputDefinition]:
return [
+ OutputDefinition(name="inference_id", kind=[STRING_KIND]),
OutputDefinition(
name="predictions", kind=[BATCH_OF_OBJECT_DETECTION_PREDICTION_KIND]
),
@@ -322,6 +325,7 @@ def _post_process_result(
predictions: List[dict],
class_filter: Optional[List[str]],
) -> BlockResult:
+ inference_id = predictions[0].get(INFERENCE_ID_KEY, None)
predictions = convert_inference_detections_batch_to_sv_detections(predictions)
predictions = attach_prediction_type_info_to_sv_detections_batch(
predictions=predictions,
@@ -335,4 +339,7 @@ def _post_process_result(
images=images,
predictions=predictions,
)
- return [{"predictions": prediction} for prediction in predictions]
+ return [
+ {"inference_id": inference_id, "predictions": prediction}
+ for prediction in predictions
+ ]
diff --git a/inference/core/workflows/core_steps/sinks/roboflow/dataset_upload/v1.py b/inference/core/workflows/core_steps/sinks/roboflow/dataset_upload/v1.py
index 14df54def0..488b6952b6 100644
--- a/inference/core/workflows/core_steps/sinks/roboflow/dataset_upload/v1.py
+++ b/inference/core/workflows/core_steps/sinks/roboflow/dataset_upload/v1.py
@@ -405,7 +405,7 @@ def execute_registration(
return False, status
except Exception as error:
credit_to_be_returned = True
- logging.exception("Failed to register datapoint at Roboflow platform")
+ logging.exception("Failed to register datapoint on the Roboflow platform")
return (
True,
f"Error while registration. Error type: {type(error)}. Details: {error}",
diff --git a/mkdocs.yml b/mkdocs.yml
index 0fb11a98b8..40c37056ed 100644
--- a/mkdocs.yml
+++ b/mkdocs.yml
@@ -68,15 +68,25 @@ nav:
- Retrieve Your API Key: quickstart/configure_api_key.md
- Model Licenses: https://roboflow.com/licensing
- Workflows:
- - What is a Workflow?: workflows/about.md
- - Understanding Workflows: workflows/understanding.md
- - Workflows definitions: workflows/definitions.md
- - How to Create and Run a Workflow: workflows/create_and_run.md
- - Blocks: workflows/blocks.md
- - Kinds: workflows/kinds.md
- - Advanced Topics:
- - Create a New Workflow Block: workflows/workflows_contribution.md
- - Modes of Running Workflows: workflows/modes_of_running.md
+ - About Workflows: workflows/about.md
+ - Create and Run: workflows/create_and_run.md
+ - Ecosystem: workflows/understanding.md
+ - Blocks overview: workflows/blocks_connections.md
+ - Blocks gallery: workflows/blocks.md
+ - Examples: workflows/gallery_index.md
+ - User guide:
+ - Running Workflows: workflows/modes_of_running.md
+ - Workflows Definitions: workflows/definitions.md
+ - Workflow execution: workflows/workflow_execution.md
+ - Developer guide:
+ - Compiler: workflows/workflows_compiler.md
+ - Execution Engine: workflows/workflows_execution_engine.md
+ - Kinds: workflows/kinds.md
+ - Blocks development: workflows/create_workflow_block.md
+ - Plugins: workflows/blocks_bundling.md
+ - Dynamic Python blocks: workflows/custom_python_code_blocks.md
+ - Versioning: workflows/versioning.md
+ - Testing: workflows/testing.md
- Reference:
- Inference Pipeline: using_inference/inference_pipeline.md
- Active Learning:
diff --git a/tests/workflows/integration_tests/execution/test_workflow_detection_plus_classification.py b/tests/workflows/integration_tests/execution/test_workflow_detection_plus_classification.py
index 77b0e6df75..07b9ddb004 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_detection_plus_classification.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_detection_plus_classification.py
@@ -5,6 +5,9 @@
from inference.core.managers.base import ModelManager
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import (
+ add_to_workflows_gallery,
+)
DETECTION_PLUS_CLASSIFICATION_WORKFLOW = {
"version": "1.0",
@@ -40,6 +43,25 @@
}
+@add_to_workflows_gallery(
+ category="Workflows with multiple models",
+ use_case_title="Workflow detection model followed by classifier",
+ use_case_description="""
+This example showcases how to stack models on top of each other - in this particular
+case, we detect objects using object detection models, requesting only "dogs" bounding boxes
+in the output of prediction.
+
+Based on the model predictions, we take each bounding box with dog and apply dynamic cropping
+to be able to run classification model for each and every instance of dog separately.
+Please note that for each inserted image we will have nested batch of crops (with size
+dynamically determined in runtime, based on first model predictions) and for each crop
+we apply secondary model.
+
+Secondary model is supposed to make prediction from dogs breed classifier model
+to assign detailed class for each dog instance.
+ """,
+ workflow_definition=DETECTION_PLUS_CLASSIFICATION_WORKFLOW,
+)
def test_detection_plus_classification_workflow_when_minimal_valid_input_provided(
model_manager: ModelManager,
dogs_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_inference_id_response.py b/tests/workflows/integration_tests/execution/test_workflow_inference_id_response.py
new file mode 100644
index 0000000000..6382386175
--- /dev/null
+++ b/tests/workflows/integration_tests/execution/test_workflow_inference_id_response.py
@@ -0,0 +1,189 @@
+import numpy as np
+import pytest
+
+from inference.core.env import WORKFLOWS_MAX_CONCURRENT_STEPS
+from inference.core.managers.base import ModelManager
+from inference.core.workflows.core_steps.common.entities import StepExecutionMode
+from inference.core.workflows.execution_engine.core import ExecutionEngine
+
+DETECTION_PLUS_CLASSIFICATION_WORKFLOW = {
+ "version": "1.0",
+ "inputs": [{"type": "WorkflowImage", "name": "image"}],
+ "steps": [
+ {
+ "type": "ObjectDetectionModel",
+ "name": "general_detection",
+ "image": "$inputs.image",
+ "model_id": "yolov8n-640",
+ "class_filter": ["dog"],
+ },
+ {
+ "type": "Crop",
+ "name": "cropping",
+ "image": "$inputs.image",
+ "predictions": "$steps.general_detection.predictions",
+ },
+ {
+ "type": "ClassificationModel",
+ "name": "breds_classification",
+ "image": "$steps.cropping.crops",
+ "model_id": "dog-breed-xpaq6/1",
+ },
+ ],
+ "outputs": [
+ {
+ "type": "JsonField",
+ "name": "predictions",
+ "selector": "$steps.breds_classification.predictions",
+ },
+ ],
+}
+
+OBJECT_DETECTION_WORKFLOW = {
+ "version": "1.0",
+ "inputs": [{"type": "WorkflowImage", "name": "image"}],
+ "steps": [
+ {
+ "type": "ObjectDetectionModel",
+ "name": "general_detection",
+ "image": "$inputs.image",
+ "model_id": "yolov8n-640",
+ "class_filter": ["dog"],
+ },
+ ],
+ "outputs": [
+ {
+ "type": "JsonField",
+ "name": "predictions",
+ "coordinates_system": "own",
+ "selector": "$steps.general_detection.predictions",
+ }
+ ],
+}
+
+INSTANCE_SEGMENTATION_WORKFLOW = {
+ "version": "1.0",
+ "inputs": [{"type": "WorkflowImage", "name": "image"}],
+ "steps": [
+ {
+ "type": "InstanceSegmentationModel",
+ "name": "instance_segmentation",
+ "image": "$inputs.image",
+ "model_id": "yolov8n-640",
+ },
+ ],
+ "outputs": [
+ {
+ "type": "JsonField",
+ "name": "predictions",
+ "selector": "$steps.instance_segmentation.*",
+ }
+ ],
+}
+
+
+@pytest.mark.workflows
+def test_detection_plus_classification_workflow_with_inference_id(
+ model_manager: ModelManager,
+ dogs_image: np.ndarray,
+ roboflow_api_key: str,
+) -> None:
+ # given
+ workflow_init_parameters = {
+ "workflows_core.model_manager": model_manager,
+ "workflows_core.api_key": roboflow_api_key,
+ "workflows_core.step_execution_mode": StepExecutionMode.LOCAL,
+ }
+ execution_engine = ExecutionEngine.init(
+ workflow_definition=DETECTION_PLUS_CLASSIFICATION_WORKFLOW,
+ init_parameters=workflow_init_parameters,
+ max_concurrent_steps=WORKFLOWS_MAX_CONCURRENT_STEPS,
+ )
+
+ # when
+ result = execution_engine.run(
+ runtime_parameters={
+ "image": dogs_image,
+ }
+ )
+
+ # then
+ assert (
+ len(result[0]["predictions"]) == 2
+ ), "Expected 2 dogs crops on input image, hence 2 nested classification results"
+
+ for prediction in result[0]["predictions"]:
+ assert "inference_id" in prediction, "Expected inference_id in each prediction"
+ assert prediction["inference_id"] is not None, "Expected non-null inference_id"
+
+ assert [result[0]["predictions"][0]["top"], result[0]["predictions"][1]["top"]] == [
+ "116.Parson_russell_terrier",
+ "131.Wirehaired_pointing_griffon",
+ ], "Expected predictions to be as measured in reference run"
+
+
+@pytest.mark.workflows
+def test_object_detection_workflow_with_inference_id(
+ model_manager: ModelManager,
+ dogs_image: np.ndarray,
+ roboflow_api_key: str,
+) -> None:
+ # given
+ workflow_init_parameters = {
+ "workflows_core.model_manager": model_manager,
+ "workflows_core.api_key": roboflow_api_key,
+ "workflows_core.step_execution_mode": StepExecutionMode.LOCAL,
+ }
+ execution_engine = ExecutionEngine.init(
+ workflow_definition=OBJECT_DETECTION_WORKFLOW,
+ init_parameters=workflow_init_parameters,
+ max_concurrent_steps=WORKFLOWS_MAX_CONCURRENT_STEPS,
+ )
+
+ # when
+ result = execution_engine.run(
+ runtime_parameters={
+ "image": dogs_image,
+ }
+ )
+
+ # then
+ assert len(result[0]["predictions"]) == 2, "Expected 2 predictions"
+ assert (
+ result[0]["predictions"][0]["inference_id"] is not None
+ ), "Expected non-null inference_id"
+ assert (
+ result[0]["predictions"][1]["inference_id"] is not None
+ ), "Expected non-null inference_id"
+
+
+@pytest.mark.workflows
+def test_instance_segmentation_workflow_with_inference_id(
+ model_manager: ModelManager,
+ dogs_image: np.ndarray,
+ roboflow_api_key: str,
+) -> None:
+ # given
+ workflow_init_parameters = {
+ "workflows_core.model_manager": model_manager,
+ "workflows_core.api_key": roboflow_api_key,
+ "workflows_core.step_execution_mode": StepExecutionMode.LOCAL,
+ }
+ execution_engine = ExecutionEngine.init(
+ workflow_definition=INSTANCE_SEGMENTATION_WORKFLOW,
+ init_parameters=workflow_init_parameters,
+ max_concurrent_steps=WORKFLOWS_MAX_CONCURRENT_STEPS,
+ )
+
+ # when
+ result = execution_engine.run(
+ runtime_parameters={
+ "image": dogs_image,
+ }
+ )
+
+ # then
+ assert len(result[0]["predictions"]) == 2, "Expected 2 predictions"
+ assert (
+ result[0]["predictions"].get("inference_id") is not None
+ ), "Expected non-null inference_id"
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_clip.py b/tests/workflows/integration_tests/execution/test_workflow_with_clip.py
index e07116b71c..4ad11b96b1 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_clip.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_clip.py
@@ -6,6 +6,9 @@
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.errors import RuntimeInputError
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import (
+ add_to_workflows_gallery,
+)
CLIP_WORKFLOW = {
"version": "1.0",
@@ -31,6 +34,19 @@
}
+@add_to_workflows_gallery(
+ category="Basic Workflows",
+ use_case_title="Workflow with CLIP model",
+ use_case_description="""
+This is the basic workflow that only contains a single CLIP model block.
+
+Please take a look at how batch-oriented WorkflowImage data is plugged to
+detection step via input selector (`$inputs.image`) and how non-batch parameters
+(reference set of texts that the each image in batch will be compared to)
+is dynamically specified - via `$inputs.reference` selector.
+ """,
+ workflow_definition=CLIP_WORKFLOW,
+)
def test_clip_workflow_when_minimal_valid_input_provided(
model_manager: ModelManager,
license_plate_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_contours_detection.py b/tests/workflows/integration_tests/execution/test_workflow_with_contours_detection.py
index 39a5fd27fd..cb79534c75 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_contours_detection.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_contours_detection.py
@@ -4,6 +4,7 @@
from inference.core.managers.base import ModelManager
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import add_to_workflows_gallery
WORKFLOW_WITH_CONTOUR_DETECTION = {
"version": "1.0",
@@ -75,18 +76,19 @@
}
+@add_to_workflows_gallery(
+ category="Workflows with classical Computer Vision methods",
+ use_case_title="Workflow detecting contours",
+ use_case_description="""
+In this example we show how classical contour detection works in cooperation
+with blocks performing its pre-processing (conversion to gray and blur).
+ """,
+ workflow_definition=WORKFLOW_WITH_CONTOUR_DETECTION,
+)
def test_workflow_with_classical_contour_detection(
model_manager: ModelManager,
dogs_image: np.ndarray,
) -> None:
- """
- In this test set we check how classical contour detection works in cooperation
- with expression block.
-
- Please point out that a single image is passed as input, and the output is a
- number of contours detected in the image.
- """
-
# given
workflow_init_parameters = {
"workflows_core.model_manager": model_manager,
@@ -105,8 +107,6 @@ def test_workflow_with_classical_contour_detection(
}
)
- print(result)
-
# then
assert isinstance(result, list), "Expected result to be list"
assert len(result) == 1, "One image provided, so one output expected"
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_counting_pixels_with_dominant_color.py b/tests/workflows/integration_tests/execution/test_workflow_with_counting_pixels_with_dominant_color.py
index cfd6792457..0595258d90 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_counting_pixels_with_dominant_color.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_counting_pixels_with_dominant_color.py
@@ -5,6 +5,7 @@
from inference.core.managers.base import ModelManager
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import add_to_workflows_gallery
WORKFLOW_WITH_PIXELS_COUNTING = {
"version": "1.0",
@@ -35,6 +36,16 @@
}
+@add_to_workflows_gallery(
+ category="Workflows with classical Computer Vision methods",
+ use_case_title="Workflow calculating pixels with dominant color",
+ use_case_description="""
+This example shows how Dominant Color block and Pixel Color Count block can be used together.
+
+First, dominant color gets detected and then number of pixels with that color is calculated.
+ """,
+ workflow_definition=WORKFLOW_WITH_PIXELS_COUNTING,
+)
def test_workflow_with_color_of_pixels_counting(
model_manager: ModelManager,
) -> None:
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_custom_python_block.py b/tests/workflows/integration_tests/execution/test_workflow_with_custom_python_block.py
index 9b9fc6c3d4..ca6a9c9d2a 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_custom_python_block.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_custom_python_block.py
@@ -12,6 +12,9 @@
)
from inference.core.workflows.execution_engine.core import ExecutionEngine
from inference.core.workflows.execution_engine.v1.dynamic_blocks import block_assembler
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import (
+ add_to_workflows_gallery,
+)
FUNCTION_TO_GET_OVERLAP_OF_BBOXES = """
def run(self, predictions: sv.Detections, class_x: str, class_y: str) -> BlockResult:
@@ -163,6 +166,27 @@ def run(self, overlaps: List[List[float]]) -> BlockResult:
}
+@add_to_workflows_gallery(
+ category="Workflows with dynamic Python Blocks",
+ use_case_title="Workflow measuring bounding boxes overlap",
+ use_case_description="""
+In real world use-cases you may not be able to find all pieces of functionalities required to complete
+your workflow within existing blocks.
+
+In such cases you may create piece of python code and put it in workflow as a dynamic block. Specifically
+here, we define two dynamic blocks:
+
+- `OverlapMeasurement` which will accept object detection predictions and provide for boxes
+of specific class matrix of overlap with all boxes of another class.
+
+- `MaximumOverlap` that will take overlap matrix produced by `OverlapMeasurement` and calculate maximum overlap.
+
+Dynamic block may be used to create steps, exactly as if those blocks were standard Workflow blocks
+existing in ecosystem. The workflow presented in the example predicts from object detection model and
+calculates overlap matrix. Later, only if more than one object is detected, maximum overlap is calculated.
+ """,
+ workflow_definition=WORKFLOW_WITH_OVERLAP_MEASUREMENT,
+)
def test_workflow_with_custom_python_blocks_measuring_overlap(
model_manager: ModelManager,
dogs_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_detection_plus_ocr.py b/tests/workflows/integration_tests/execution/test_workflow_with_detection_plus_ocr.py
index 067b328ea9..ec1c023a67 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_detection_plus_ocr.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_detection_plus_ocr.py
@@ -5,6 +5,9 @@
from inference.core.managers.base import ModelManager
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import (
+ add_to_workflows_gallery,
+)
MULTI_STAGES_WORKFLOW = {
"version": "1.0",
@@ -77,6 +80,28 @@
}
+@add_to_workflows_gallery(
+ category="Workflows with multiple models",
+ use_case_title="Workflow detection models and OCR",
+ use_case_description="""
+This example showcases quite sophisticated workflows usage scenario that assume the following:
+
+- we have generic object detection model capable of recognising cars
+
+- we have specialised object detection model trained to detect license plates in the images depicting **single car only**
+
+- we have generic OCR model capable of recognising lines of texts from images
+
+Our goal is to read license plates of every car we detect in the picture. We can achieve that goal with
+workflow from this example. In the definition we can see that generic object detection model is applied first,
+to make the job easier for the secondary (plates detection) model we enlarge bounding boxes, slightly
+offsetting its dimensions with Detections Offset block - later we apply cropping to be able to run
+license plate detection for every detected car instance (increasing the depth of the batch). Once secondary model
+runs and we have bounding boxes for license plates - we crop previously cropped cars images to extract plates.
+Once this is done, plates crops are passed to OCR step which turns images of plates into text.
+""",
+ workflow_definition=MULTI_STAGES_WORKFLOW,
+)
def test_detection_plus_ocr_workflow_when_minimal_valid_input_provided(
model_manager: ModelManager,
license_plate_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_detections_classes_replacement_by_specialised_classification.py b/tests/workflows/integration_tests/execution/test_workflow_with_detections_classes_replacement_by_specialised_classification.py
index fe3e8ec23a..5529cff5a4 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_detections_classes_replacement_by_specialised_classification.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_detections_classes_replacement_by_specialised_classification.py
@@ -5,6 +5,9 @@
from inference.core.managers.base import ModelManager
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import (
+ add_to_workflows_gallery,
+)
DETECTION_CLASSES_REPLACEMENT_WORKFLOW = {
"version": "1.0",
@@ -51,6 +54,27 @@
}
+@add_to_workflows_gallery(
+ category="Workflows with multiple models",
+ use_case_title="Workflow with classifier providing detailed labels for detected objects",
+ use_case_description="""
+This example illustrates how helpful Workflows could be when you have generic object detection model
+(capable of detecting common classes - like dogs) and specific classifier (capable of providing granular
+predictions for narrow high-level classes of objects - like dogs breed classifier). Having list
+of classifier predictions for each detected dog is not handy way of dealing with output -
+as you kind of loose the information about location of specific dog. To avoid this problem, you
+may want to replace class labels of original bounding boxes (from the first model localising dogs) with
+classes predicted by classifier.
+
+In this example, we use Detections Classes Replacement block which is also interesting from the
+perspective of difference of its inputs dimensionality levels. `object_detection_predictions` input
+has level 1 (there is one prediction with bboxes for each input image) and `classification_predictions`
+has level 2 (there are bunch of classification results for each input image). The block combines that
+two inputs and produces result at dimensionality level 1 - exactly the same as predictions from
+object detection model.
+ """,
+ workflow_definition=DETECTION_CLASSES_REPLACEMENT_WORKFLOW,
+)
def test_detection_plus_classification_workflow_when_minimal_valid_input_provided(
model_manager: ModelManager,
dogs_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_detections_consensus_block.py b/tests/workflows/integration_tests/execution/test_workflow_with_detections_consensus_block.py
index f5d516925e..720f4d5233 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_detections_consensus_block.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_detections_consensus_block.py
@@ -7,6 +7,9 @@
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.errors import RuntimeInputError, StepExecutionError
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import (
+ add_to_workflows_gallery,
+)
CONSENSUS_WORKFLOW = {
"version": "1.0",
@@ -61,6 +64,25 @@
)
+@add_to_workflows_gallery(
+ category="Workflows with multiple models",
+ use_case_title="Workflow presenting models ensemble",
+ use_case_description="""
+This workflow presents how to combine predictions from multiple models running against the same
+input image with the block called Detections Consensus.
+
+First, we run two object detections models steps and we combine their predictions. Fusion may be
+performed in different scenarios based on Detections Consensus step configuration:
+
+- you may combine predictions from models detecting different objects and then require only single
+model vote to add predicted bounding box to the output prediction
+
+- you may combine predictions from models detecting the same objects and expect multiple positive
+votes to accept bounding box to the output prediction - this way you may improve the quality of
+predictions
+ """,
+ workflow_definition=CONSENSUS_WORKFLOW,
+)
def test_consensus_workflow_when_minimal_valid_input_provided(
model_manager: ModelManager,
crowd_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_dominant_color.py b/tests/workflows/integration_tests/execution/test_workflow_with_dominant_color.py
index 753a563a2f..5141a327cd 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_dominant_color.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_dominant_color.py
@@ -4,6 +4,7 @@
from inference.core.managers.base import ModelManager
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import add_to_workflows_gallery
MINIMAL_DOMINANT_COLOR_WORKFLOW = {
"version": "1.0",
@@ -26,6 +27,14 @@
}
+@add_to_workflows_gallery(
+ category="Workflows with classical Computer Vision methods",
+ use_case_title="Workflow calculating dominant color",
+ use_case_description="""
+This example shows how Dominant Color block can be used against input image.
+ """,
+ workflow_definition=MINIMAL_DOMINANT_COLOR_WORKFLOW,
+)
def test_dominant_color_workflow_when_minimal_valid_input_provided(
model_manager: ModelManager,
red_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_filtering.py b/tests/workflows/integration_tests/execution/test_workflow_with_filtering.py
index efce671a64..c3098ddd39 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_filtering.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_filtering.py
@@ -10,6 +10,9 @@
)
from inference.core.workflows.errors import RuntimeInputError, StepExecutionError
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import (
+ add_to_workflows_gallery,
+)
FILTERING_WORKFLOW = {
"version": "1.0",
@@ -108,6 +111,19 @@
)
+@add_to_workflows_gallery(
+ category="Workflows with data transformations",
+ use_case_title="Workflow with detections filtering",
+ use_case_description="""
+This example presents how to use Detections Transformation block to build workflow
+that is going to filter predictions based on:
+
+- predicted classes
+
+- size of predicted bounding box relative to size of input image
+ """,
+ workflow_definition=FILTERING_WORKFLOW,
+)
def test_filtering_workflow_when_minimal_valid_input_provided(
model_manager: ModelManager,
crowd_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_flow_control.py b/tests/workflows/integration_tests/execution/test_workflow_with_flow_control.py
index ffe7aeb81d..c59daef2dd 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_flow_control.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_flow_control.py
@@ -9,6 +9,9 @@
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.execution_engine.core import ExecutionEngine
from inference.core.workflows.execution_engine.introspection import blocks_loader
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import (
+ add_to_workflows_gallery,
+)
AB_TEST_WORKFLOW = {
"version": "1.0",
@@ -413,29 +416,26 @@ def test_flow_control_step_affecting_batches(
}
+@add_to_workflows_gallery(
+ category="Workflows with flow control",
+ use_case_title="Workflow with if statement applied on nested batches",
+ use_case_description="""
+In this test scenario we verify if we can successfully apply conditional
+branching when data dimensionality increases.
+We first make detections on input images and perform crop increasing
+dimensionality to 2. Then we make another detections on cropped images
+and check if inside crop we only see one instance of class dog (very naive
+way of making sure that bboxes contain only single objects).
+Only if that condition is true, we run classification model - to
+classify dog breed.
+ """,
+ workflow_definition=WORKFLOW_WITH_CONDITION_DEPENDENT_ON_CROPS,
+)
def test_flow_control_step_affecting_data_with_increased_dimensionality(
model_manager: ModelManager,
crowd_image: np.ndarray,
dogs_image: np.ndarray,
) -> None:
- """
- In this test scenario we verify if we can successfully apply conditional
- branching when data dimensionality increases.
- We first make detections on input images and perform crop increasing
- dimensionality to 2. Then we make another detections on cropped images
- and check if inside crop we only see one instance of class dog (very naive
- way of making sure that bboxes contain only single objects).
- Only if that condition is true, we run classification model - to
- classify dog breed.
-
- What is verified from EE standpoint:
- * Creating execution branches for each batch element separately on deeper
- dimensionality levels and then executing downstream step according to
- decision made previously - with execution branches being independent
- * proper behavior of steps expecting non-empty inputs w.r.t. masks for
- execution branches
- * correctness of building nested outputs
- """
# given
workflow_init_parameters = {
"workflows_core.model_manager": model_manager,
@@ -564,6 +564,18 @@ def test_flow_control_step_affecting_data_with_increased_dimensionality(
}
+@add_to_workflows_gallery(
+ category="Workflows with flow control",
+ use_case_title="Workflow with if statement applied on non batch-oriented input",
+ use_case_description="""
+In this test scenario we show that we can use non-batch oriented conditioning (ContinueIf block).
+
+If statement is effectively applied on input parameter that would determine path of execution for
+all data passed in `image` input. When the value matches expectation - all dependent steps
+will be executed, otherwise only the independent ones.
+ """,
+ workflow_definition=WORKFLOW_WITH_CONDITION_DEPENDENT_ON_CROPS,
+)
def test_flow_control_workflow_where_non_batch_nested_parameter_affects_further_execution_when_condition_is_met(
model_manager: ModelManager,
crowd_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_model_running_on_absolute_static_crop.py b/tests/workflows/integration_tests/execution/test_workflow_with_model_running_on_absolute_static_crop.py
index 06b1e833a0..4621b22f21 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_model_running_on_absolute_static_crop.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_model_running_on_absolute_static_crop.py
@@ -7,6 +7,9 @@
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.errors import RuntimeInputError
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import (
+ add_to_workflows_gallery,
+)
ABSOLUTE_STATIC_CROP_WORKFLOW = {
"version": "1.0",
@@ -50,6 +53,21 @@
}
+@add_to_workflows_gallery(
+ category="Basic Workflows",
+ use_case_title="Workflow with static crop and object detection model",
+ use_case_description="""
+This is the basic workflow that contains single transformation (static crop)
+followed by object detection model. This example may be inspiration for anyone
+who would like to run specific model only on specific part of the image.
+The Region of Interest does not necessarily have to be defined statically -
+please note that coordinates of static crops are referred via input selectors,
+which means that each time you run the workflow (for instance in each different
+physical location, where RoI for static crop is location-dependent) you may
+provide different RoI coordinates.
+ """,
+ workflow_definition=ABSOLUTE_STATIC_CROP_WORKFLOW,
+)
def test_static_crop_workflow_when_minimal_valid_input_provided(
model_manager: ModelManager,
crowd_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_property_extraction.py b/tests/workflows/integration_tests/execution/test_workflow_with_property_extraction.py
index 15e6dcfd55..bad0c91a78 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_property_extraction.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_property_extraction.py
@@ -6,6 +6,9 @@
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.errors import StepOutputLineageError
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import (
+ add_to_workflows_gallery,
+)
WORKFLOW_WITH_EXTRACTION_OF_CLASSES_FOR_DETECTIONS = {
"version": "1.0",
@@ -90,6 +93,40 @@
}
+@add_to_workflows_gallery(
+ category="Workflows with business logic",
+ use_case_title="Workflow with extraction of classes for detections (1)",
+ use_case_description="""
+In practical use-cases you may find the need to inject pieces of business logic inside
+your Workflow, such that it is easier to integrate with app created in Workflows ecosystem.
+
+Translation of model predictions into domain-specific language of your business is possible
+with specialised blocks that let you parametrise such programming constructs
+as switch-case statements.
+
+In this example, our goal is to:
+
+- tell how many objects are detected
+
+- verify that the picture presents exactly two dogs
+
+To achieve that goal, we run generic object detection model as first step, then we use special
+block called Property Definition that is capable of executing various operations to
+transform input data into desired output. We have two such blocks:
+
+- `instances_counter` which takes object detection predictions and apply operation to extract sequence length -
+effectively calculating number of instances of objects that were predicted
+
+- `property_extraction` which extracts class names from all detected bounding boxes
+
+`instances_counter` basically completes first goal of the workflow, but to satisfy the second one we need to
+build evaluation logic that will tell "PASS" / "FAIL" based on comparison of extracted class names with
+reference parameter (provided via Workflow input `$inputs.reference`). We can use Expression block to achieve
+that goal - building custom case statements (checking if class names being list of classes
+extracted from object detection prediction matches reference passed in the input).
+ """,
+ workflow_definition=WORKFLOW_WITH_EXTRACTION_OF_CLASSES_FOR_DETECTIONS,
+)
def test_workflow_with_extraction_of_classes_for_detections(
model_manager: ModelManager,
dogs_image: np.ndarray,
@@ -250,6 +287,55 @@ def test_workflow_with_extraction_of_classes_for_detections(
}
+@add_to_workflows_gallery(
+ category="Workflows with business logic",
+ use_case_title="Workflow with extraction of classes for detections (2)",
+ use_case_description="""
+In practical use-cases you may find the need to inject pieces of business logic inside
+your Workflow, such that it is easier to integrate with app created in Workflows ecosystem.
+
+Translation of model predictions into domain-specific language of your business is possible
+with specialised blocks that let you parametrise such programming constructs
+as switch-case statements.
+
+In this example, our goal is to:
+
+- run generic object detection model to find instances of dogs
+
+- crop dogs detection
+
+- run specialised dogs breed classifier to assign granular label for each dog
+
+- compare predicted dogs breeds to verify if detected labels matches exactly reverence value passed in input.
+
+This example is quite complex as it requires quite deep understanding of Workflows ecosystem. Let's start from
+the beginning - we run object detection model, crop its detections according to dogs class to perform
+classification. This is quite typical for workflows (you may find such pattern in remaining examples).
+
+The complexity increases when we try to handle classification output. We need to have a list of classes
+for each input image, but for now we have complex objects with all classification predictions details
+provided by `breds_classification` step - what is more - we have batch of such predictions for
+each input image (as we created dogs crops based on object detection model predictions). To solve the
+problem, at first we apply Property Definition step taking classifier predictions and turning them into
+strings representing predicted classes. We still have batch of class names at dimensionality level 2,
+which needs to be brought into dimensionality level 1 to make a single comparison against reference
+value for each input image. To achieve that effect we use Dimension Collapse block which does nothing
+else but grabs the batch of classes and turns it into list of classes at dimensionality level 1 - one
+list for each input image.
+
+That would solve our problems, apart from one nuance that must be taken into account. First-stage model
+is not guaranteed to detect any dogs - and if that happens we do not execute cropping and further
+processing for that image, leaving all outputs derived from downstream computations `None` which is
+suboptimal. To compensate for that, we may use First Non Empty Or Default block which will take
+`outputs_concatenation` step output and replace missing values with empty list (as effectively this is
+equivalent of not detecting any dog).
+
+Such prepared output of `empty_values_replacement` step may be now plugged into Expression block,
+performing switch-case like logic to deduce if breeds of detected dogs match with reference value
+passed to workflow execution.
+ """,
+ workflow_definition=WORKFLOW_WITH_EXTRACTION_OF_CLASS_NAME_FROM_CROPS_AND_CONCATENATION_OF_RESULTS,
+)
def test_workflow_with_extraction_of_classes_for_classification_on_crops(
model_manager: ModelManager,
dogs_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_sahi.py b/tests/workflows/integration_tests/execution/test_workflow_with_sahi.py
index b5ab332bf9..41494a1758 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_sahi.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_sahi.py
@@ -7,6 +7,7 @@
from inference.core.managers.base import ModelManager
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import add_to_workflows_gallery
SAHI_WORKFLOW = {
"version": "1.0.0",
@@ -56,6 +57,24 @@
}
+@add_to_workflows_gallery(
+ category="Advanced inference techniques",
+ use_case_title="SAHI in workflows - object detection",
+ use_case_description="""
+This example illustrates usage of [SAHI](https://blog.roboflow.com/how-to-use-sahi-to-detect-small-objects/)
+technique in workflows.
+
+Workflows implementation requires three blocks:
+
+- Image Slicer - which runs a sliding window over image and for each image prepares batch of crops
+
+- detection model block (in our scenario Roboflow Object Detection model) - which is responsible
+for making predictions on each crop
+
+- Detections stitch - which combines partial predictions for each slice of the image into a single prediction
+ """,
+ workflow_definition=SAHI_WORKFLOW,
+)
def test_sahi_workflow_with_none_as_filtering_strategy(
model_manager: ModelManager,
license_plate_image: np.ndarray,
@@ -408,6 +427,24 @@ def slicer_callback(image_slice: np.ndarray):
}
+@add_to_workflows_gallery(
+ category="Advanced inference techniques",
+ use_case_title="SAHI in workflows - instance segmentation",
+ use_case_description="""
+This example illustrates usage of [SAHI](https://blog.roboflow.com/how-to-use-sahi-to-detect-small-objects/)
+technique in workflows.
+
+Workflows implementation requires three blocks:
+
+- Image Slicer - which runs a sliding window over image and for each image prepares batch of crops
+
+- detection model block (in our scenario Roboflow Instance Segmentation model) - which is responsible
+for making predictions on each crop
+
+- Detections stitch - which combines partial predictions for each slice of the image into a single prediction
+ """,
+ workflow_definition=SAHI_WORKFLOW,
+)
def test_sahi_workflow_for_segmentation_with_nms_as_filtering_strategy(
model_manager: ModelManager,
license_plate_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_sam2.py b/tests/workflows/integration_tests/execution/test_workflow_with_sam2.py
index 1bc929de7d..25998df2a9 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_sam2.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_sam2.py
@@ -11,6 +11,7 @@
BlockManifest,
)
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import add_to_workflows_gallery
@pytest.mark.parametrize("images_field_alias", ["images", "image"])
@@ -85,6 +86,17 @@ def test_sam2_model_validation_when_parameters_have_invalid_type(
}
+@add_to_workflows_gallery(
+ category="Workflows with foundation models",
+ use_case_title="Workflow with Segment Anything 2 model",
+ use_case_description="""
+Meta AI introduced very capable segmentation model called [SAM 2](https://ai.meta.com/sam2/) which
+has capabilities of producing segmentation masks for instances of objects.
+
+It can be used within workflows in couple of ways -
+ """,
+ workflow_definition=SIMPLE_SAM_WORKFLOW,
+)
def test_sam2_workflow_when_minimal_valid_input_provided(
model_manager: ModelManager, dogs_image: np.ndarray
) -> None:
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_sift.py b/tests/workflows/integration_tests/execution/test_workflow_with_sift.py
index 7870d48cdf..a33e364680 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_sift.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_sift.py
@@ -4,6 +4,7 @@
from inference.core.managers.base import ModelManager
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import add_to_workflows_gallery
WORKFLOW_WITH_SIFT = {
"version": "1.0",
@@ -77,22 +78,33 @@
}
+@add_to_workflows_gallery(
+ category="Workflows with classical Computer Vision methods",
+ use_case_title="SIFT in Workflows",
+ use_case_description="""
+In this example we check how SIFT-based pattern matching works in cooperation
+with expression block.
+
+The Workflow first calculates SIFT features for input image and reference template,
+then image features are compared to template features. At the end - switch-case
+statement is built with Expression block to produce output.
+
+Important detail: If there is empty output from SIFT descriptors calculation
+for (which is a valid output if no feature gets recognised) the sift comparison won't
+execute - hence First Non Empty Or Default block is used to provide default outcome
+for `images_match` output of SIFT comparison block.
+
+Please note that a single image can be passed as template, and batch of images
+are passed as images to look for template. This workflow does also validate
+Execution Engine capabilities to broadcast batch-oriented inputs properly.
+ """,
+ workflow_definition=WORKFLOW_WITH_SIFT,
+)
def test_workflow_with_classical_pattern_matching(
model_manager: ModelManager,
dogs_image: np.ndarray,
crowd_image: np.ndarray,
) -> None:
- """
- In this test set we check how SIFT-based pattern matching works in cooperation
- with expression block.
-
- Please point out that a single image is passed as template, and batch of images
- are passed as images to look for template. This workflow does also validate
- Execution Engine capabilities to broadcast batch-oriented inputs properly.
-
- Additionally, there is empty output from SIFT descriptors calculation
- for blank input, which is nicely handled by first_non_empty_or_default block.
- """
# given
template = np.ascontiguousarray(dogs_image[::-1, ::-1], dtype=np.uint8)
empty_image_without_descriptors = np.zeros((256, 256, 3), dtype=np.uint8)
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_single_model.py b/tests/workflows/integration_tests/execution/test_workflow_with_single_model.py
index 0c9a256643..c36b8c243e 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_single_model.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_single_model.py
@@ -7,6 +7,9 @@
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.errors import RuntimeInputError, StepExecutionError
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import (
+ add_to_workflows_gallery,
+)
OBJECT_DETECTION_WORKFLOW = {
"version": "1.0",
@@ -63,6 +66,18 @@
)
+@add_to_workflows_gallery(
+ category="Basic Workflows",
+ use_case_title="Workflow with single object detection model",
+ use_case_description="""
+This is the basic workflow that only contains a single object detection model.
+
+Please take a look at how batch-oriented WorkflowImage data is plugged to
+detection step via input selector (`$inputs.image`) and how non-batch parameters
+are dynamically specified - via `$inputs.model_id` and `$inputs.confidence` selectors.
+ """,
+ workflow_definition=OBJECT_DETECTION_WORKFLOW,
+)
def test_object_detection_workflow_when_minimal_valid_input_provided(
model_manager: ModelManager,
crowd_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_sorting.py b/tests/workflows/integration_tests/execution/test_workflow_with_sorting.py
index a92d6eec4e..0be78c3630 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_sorting.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_sorting.py
@@ -13,6 +13,9 @@
)
from inference.core.workflows.errors import RuntimeInputError, StepExecutionError
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import (
+ add_to_workflows_gallery,
+)
def build_sorting_workflow_definition(
@@ -95,6 +98,19 @@ def test_sorting_workflow_for_when_nothing_to_sort(
assert len(detections) == 0, "Expected nothing to pass confidence threshold"
+@add_to_workflows_gallery(
+ category="Workflows with data transformations",
+ use_case_title="Workflow with detections sorting",
+ use_case_description="""
+This workflow presents how to use Detections Transformation block that is going to
+align predictions from object detection model such that results are sorted
+ascending regarding confidence.
+ """,
+ workflow_definition=build_sorting_workflow_definition(
+ sort_operation_mode=DetectionsSortProperties.CONFIDENCE,
+ ascending=True,
+ ),
+)
def test_sorting_workflow_for_confidence_ascending(
model_manager: ModelManager,
crowd_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/test_workflow_with_stitch_for_dynamic_crop.py b/tests/workflows/integration_tests/execution/test_workflow_with_stitch_for_dynamic_crop.py
index 8ece373e43..e33512d077 100644
--- a/tests/workflows/integration_tests/execution/test_workflow_with_stitch_for_dynamic_crop.py
+++ b/tests/workflows/integration_tests/execution/test_workflow_with_stitch_for_dynamic_crop.py
@@ -4,6 +4,7 @@
from inference.core.managers.base import ModelManager
from inference.core.workflows.core_steps.common.entities import StepExecutionMode
from inference.core.workflows.execution_engine.core import ExecutionEngine
+from tests.workflows.integration_tests.execution.workflows_gallery_collector.decorators import add_to_workflows_gallery
WORKFLOW_WITH_DYNAMIC_CROP_AND_STITCH = {
"version": "1.0.0",
@@ -68,6 +69,28 @@
}
+@add_to_workflows_gallery(
+ category="Workflows with visualization blocks",
+ use_case_title="Predictions from different models visualised together",
+ use_case_description="""
+This workflow showcases how predictions from different models (even from nested
+batches created from input images) may be visualised together.
+
+Our scenario covers:
+
+- Detecting cars using YOLOv8 model
+
+- Dynamically cropping input images to run secondary model (license plates detector) for each
+car instance
+
+- Stitching together all predictions for licence plates into single prediction
+
+- Fusing cars detections and license plates detections into single prediction
+
+- Visualizing final predictions
+ """,
+ workflow_definition=WORKFLOW_WITH_DYNAMIC_CROP_AND_STITCH,
+)
def test_workflow_with_stitch_and_dynamic_crop(
model_manager: ModelManager,
license_plate_image: np.ndarray,
diff --git a/tests/workflows/integration_tests/execution/workflows_gallery_collector/__init__.py b/tests/workflows/integration_tests/execution/workflows_gallery_collector/__init__.py
new file mode 100644
index 0000000000..e69de29bb2
diff --git a/tests/workflows/integration_tests/execution/workflows_gallery_collector/decorators.py b/tests/workflows/integration_tests/execution/workflows_gallery_collector/decorators.py
new file mode 100644
index 0000000000..f94f5f373f
--- /dev/null
+++ b/tests/workflows/integration_tests/execution/workflows_gallery_collector/decorators.py
@@ -0,0 +1,39 @@
+import functools
+from dataclasses import dataclass
+from typing import List
+
+
+@dataclass(frozen=True)
+class WorkflowGalleryEntry:
+ category: str
+ use_case_title: str
+ use_case_description: str
+ workflow_definition: dict
+
+
+GALLERY_ENTRIES: List[WorkflowGalleryEntry] = []
+
+
+def add_to_workflows_gallery(
+ category: str,
+ use_case_title: str,
+ use_case_description: str,
+ workflow_definition: dict,
+):
+ global GALLERY_ENTRIES
+ gallery_entry = WorkflowGalleryEntry(
+ category=category,
+ use_case_title=use_case_title,
+ use_case_description=use_case_description.strip(),
+ workflow_definition=workflow_definition,
+ )
+ GALLERY_ENTRIES.append(gallery_entry)
+
+ def decorator(function):
+ @functools.wraps(function)
+ def wrapper(*args, **kwargs):
+ return function(*args, **kwargs)
+
+ return wrapper
+
+ return decorator