-
Notifications
You must be signed in to change notification settings - Fork 0
/
Copy pathpart3.Rmd
499 lines (346 loc) · 19.4 KB
/
part3.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
---
title: "R Crash Course"
author: "Mark Dunning"
date: '`r format(Sys.time(), "Last modified: %d %b %Y")`'
output:
html_notebook:
toc: yes
toc_float: yes
css: stylesheets/styles.css
editor_options:
chunk_output_type: inline
---
# Introduction to R - Part III
<img src="images/logo-sm.png" style="position:absolute;top:40px;right:10px;" width="200"/>
Lets make sure we have read the gapminder data into R and have the relevant packages loaded.
```{r message=FALSE,warning=FALSE}
library(readr)
library(dplyr)
library(ggplot2)
gapminder <- read_csv("raw_data/gapminder.csv")
```
## Customising a plot
Now make a scatter plot of gdp versus life expectancy as we did in the previous session. One of the last topics we covered was how to add colour to a plot. This can make the plot more appealing, but also help with data interpretation. In this case, we can use different colours to indicate countries belonging to different continents.
```{r}
ggplot(gapminder, aes(x = gdpPercap, y=lifeExp,col=continent)) + geom_point()
```
The shape and size of points can also be mapped from the data. However, it is easy to get carried away.
```{r}
ggplot(gapminder, aes(x = gdpPercap, y=lifeExp,shape=continent,size=pop)) + geom_point()
```
Scales and their legends have so far been handled using `ggplot2` defaults. `ggplot2` offers functionality to have finer control over scales and legends using the scale methods.
Scale methods are divided into functions by combinations of
- the aesthetics they control.
- the type of data mapped to scale.
`scale_`*aesthetic*\_*type*
Try typing in `scale_` then tab to autocomplete. This will provide some examples of the scale functions available in `ggplot2`.
Although different scale functions accept some variety in their arguments, common arguments to scale functions include -
- name - The axis or legend title
- limits - Minimum and maximum of the scale
- breaks - Label/tick positions along an axis
- labels - Label names at each break
- values - the set of aesthetic values to map data values
We can choose specific colour palettes, such as those provided by the `RColorBrewer` package. This package provides palettes for different types of scale (sequential, diverging, qualitative).
```{r}
library(RColorBrewer)
display.brewer.all(colorblindFriendly = TRUE)
```
::: warning
**When creating a plot, always check that the colour scheme is appropriate for people with various forms of colour-blindness**
:::
When experimenting with colour palettes and labels, it is useful to save the plot as an object
```{r}
p <- ggplot(gapminder, aes(x = gdpPercap, y=lifeExp,col=continent)) + geom_point()
```
```{r}
# We can also change the text displayed above the legend with the name parameter.
p + scale_color_manual(values=brewer.pal(6,"Set2"))
```
Or we can even specify our own colours; such as The University of Sheffield branding colours
```{r}
uos_colours <- c("#440099","#9ADBE8","#131E29",
"#005A8F",
"#00BBCC",
"#64CBE8",
"#00CE7C",
"#3BD4AE",
"#A1DED2",
"#663DB3",
"#981F92",
"#DAA8E2",
"#E7004C",
"#FF6371",
"#FF9664"
)
uos_colours <- as.character(uos_colours)
p + scale_color_manual(values=uos_colours)
```
**NEW**:- A set of palettes based on works in the Metropolitan Museum of Art (New York) has been made available.
<https://github.com/BlakeRMills/MetBrewer>
```{r}
## this will check if MetBrewer is already installed, and will only install if it is not found
if(!require("MetBrewer")) install.packages("MetBrewer")
library(MetBrewer)
p + scale_color_manual(values=met.brewer(name = "Greek"))
```
Various labels can be modified using the `labs` function.
```{r}
p + labs(x="Wealth",y="Life Expectancy",title="Relationship between Wealth and Life Expectancy")
```
We can also modify the x- and y- limits of the plot so that any outliers are not shown. `ggplot2` will give a warning that some points are excluded.
```{r}
p + xlim(0,60000)
```
Saving is supported by the `ggsave` function. A variety of file formats are supported (`.png`, `.pdf`, `.tiff`, etc) and the format used is determined from the extension given in the `file` argument. The height, width and resolution can also be configured. See the help on `ggsave` (`?ggsave`) for more information.
```{r}
ggsave(p, file="my_ggplot.png")
```
Most aspects of the plot can be modified from the background colour to the grid sizes and font. Several pre-defined "themes" exist and we can modify the appearance of the whole plot using a `theme_..` function.
```{r}
p + theme_bw()
```
More themes are supported by the `ggthemes` package. You can make your plots look like the Economist, Wall Street Journal or Excel (**but please don't do this!**)
```{r}
## this will check if ggthemes is already installed, and will only install if it is not found
if(!require("ggthemes")) install.packages("ggthemes")
library(ggthemes)
p + theme_excel()
```
## Exercise
::: exercise
**Exercise**: Use a boxplot to compare the life expectancy values of Australia and New Zealand. Use a `Set2` palette from `RColorBrewer` to colour the boxplots and apply a "minimal" theme to the plot.
:::

## Facets
One very useful feature of `ggplot2` is faceting. This allows you to produce plots subset by variables in your data. In the scatter plot above, it was quite difficult to see if the relationship between gdp and life expectancy was the same for each continent. To overcome this, we would like a see a separate plot for each continent.
To facet our data into multiple plots we can use the `facet_wrap` (1 variable) or `facet_grid` (2 variables) functions and specify the variable(s) we split by.
```{r}
p + facet_wrap(~continent)
```
The `facet_grid` function will create a grid-like plot with one variable on the x-axis and another on the y-axis.
```{r fig.width=12}
p + facet_grid(continent~year)
```
The previous plot was a bit messy as it contained all combinations of year and continent. Let's suppose we want our analysis to be a bit more focused and disregard countries in Oceania (as there are only 2 in our dataset) and maybe years between 1997 and 2002.
We should know how to restrict the rows from the `gapminder` dataset using the `filter` function. Instead of filtering the data, creating a new data frame and constructing the data frame from these new data we can use the`%>%` operator to create the data frame on the fly and pass directly to `ggplot`. Thus we don't have to save a new data frame or alter the original data.
```{r fig.width=12}
filter(gapminder, continent!="Oceania", year %in% c(1997,2002,2007)) %>%
ggplot(aes(x = gdpPercap, y=lifeExp,col=continent)) + geom_point() + facet_grid(continent~year)
```
# Summarising and grouping with dplyr
The `summarise` function can take any R function that takes a vector of values (i.e. a column from a data frame) and returns a single value. Some of the more useful functions include:
- `min` minimum value
- `max` maximum value
- `sum` sum of values
- `mean` mean value
- `sd` standard deviation
- `median` median value
- `IQR` the interquartile range
- `n_distinct` the number of distinct values
- `n` the number of observations (Note: this is a special function that doesn't take a vector argument, i.e. column)
```{r}
library(dplyr)
summarise(gapminder, min(lifeExp), max(gdpPercap), mean(pop))
```
It is also possible to summarise using a function that takes more than one value, i.e. from multiple columns. For example, we could compute the correlation between year and life expectancy. Here we also assign names to the table that is produced.
```{r}
gapminder %>%
summarise(MinLifeExpectancy = min(lifeExp),
MaximumGDP = max(gdpPercap),
AveragePop = mean(pop),
Correlation = cor(year, lifeExp))
```
However, it is not particularly useful to calculate such values from the entire table as we have different continents and years. The `group_by` function allows us to split the table into different categories, and compute summary statistics for each year (for example).
```{r}
gapminder %>%
group_by(year) %>%
summarise(MinLifeExpectancy = min(lifeExp),
MaximumGDP = max(gdpPercap),
AveragePop = mean(pop))
```
```{r}
gapminder %>%
group_by(year,continent) %>%
summarise(MinLifeExpectancy = min(lifeExp),
MaximumGDP = max(gdpPercap),
AveragePop = mean(pop))
```
We can list as many summary functions as we like. Whilst this can make our code somewhat verbose there are many helper functions available. Consider an example where we want to average all the columns in our data:-
```{r}
gapminder %>%
group_by(year) %>%
summarise(MeanLifeExpectancy = mean(lifeExp),
MeanGDP = mean(gdpPercap),
MeanPop = mean(pop))
```
This wasn't a huge effort to write this code. However, it would be much more tedious for a dataset with many more columns. Recognising this, we can use the convenient `summarise_all` function. This will return `NA` values for columns that do not contain numeric values.
```{r warning=FALSE,message=FALSE}
gapminder %>%
group_by(continent) %>%
summarise_all(mean)
```
The nice thing about `summarise` is that it can followed up by any of the other `dplyr` verbs that we have met so far (`select`, `filter`, `arrange`..etc). As the `country` column of the previous output containing missing values we can exclude it from further processing.
```{r warning=FALSE,message=FALSE}
gapminder %>%
group_by(continent) %>%
summarise_all(mean) %>%
select(-country)
```
Returning to the correlation between life expectancy and year, we can summarise as follows:-
```{r,message=FALSE}
gapminder %>%
group_by(country) %>%
summarise(Correlation = cor(year , lifeExp))
```
We can then arrange the table by the correlation to see which countries have the lowest correlation
```{r,message=FALSE}
gapminder %>%
group_by(country) %>%
summarise(Correlation = cor(year , lifeExp)) %>%
arrange(Correlation)
```
We can filter the results to find observations of interest
```{r,message=FALSE}
gapminder %>%
group_by(country) %>%
summarise(Correlation = cor(year , lifeExp)) %>%
filter(Correlation < 0)
```
The countries we identify could then be used as the basis for a plot.
```{r}
library(ggplot2)
filter(gapminder, country %in% c("Rwanda","Zambia","Zimbabwe")) %>%
ggplot(aes(x=year, y=lifeExp,col=country)) + geom_line()
```
------------------------------------------------------------------------
------------------------------------------------------------------------
------------------------------------------------------------------------
### Exercise
::: exercise
- Summarise the `gapminder` data in an appropriate manner to produce a plot to show the change in average `gdpPercap` for each continent over time.
- see below for a suggestion
- HINT: you will need to use the `geom_col` function to create the bar plot
:::
------------------------------------------------------------------------
------------------------------------------------------------------------
------------------------------------------------------------------------
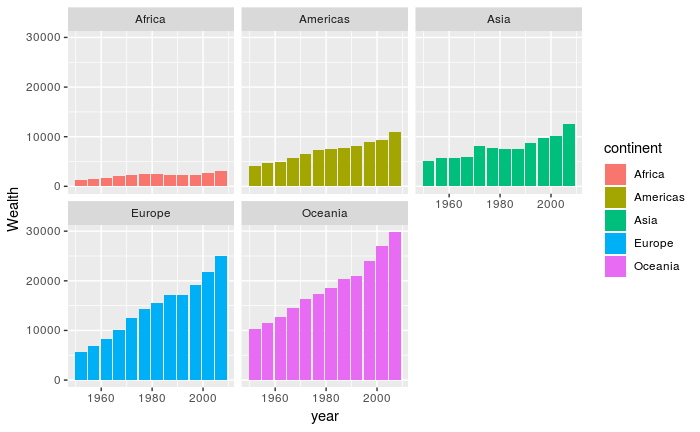
# Joining
In many real life situations, data are spread across multiple tables or spreadsheets. Usually this occurs because different types of information about a subject, e.g. a patient, are collected from different sources. It may be desirable for some analyses to combine data from two or more tables into a single data frame based on a common column, for example, an attribute that uniquely identifies the subject.
`dplyr` provides a set of join functions for combining two data frames based on matches within specified columns. For those familiar with such SQL, these operations are very similar to carrying out join operations between tables in a relational database.
As a toy example, lets consider two data frames that some results of testing whether genes A, B and C are significant in our study (gene expression, mutations, etc.)
```{r}
gene_results <- data.frame(Name=LETTERS[1:3], pvalue = c(0.001, 0.1,0.01))
gene_results
```
We might also have a data frame containing more data about the genes; such as which chromosome they are located on. As part of our data interpretation we might need to know where in the genome the genes are located. Note that both data frames have a column called `Name`. This column will be used to identify genes common to both tables.
```{r}
gene_anno <- data.frame(Name = c("A","B","D"), chromosome=c(1,1,3))
gene_anno
```
There are various ways in which we can join these two tables together. We will first consider the case of a "left join".
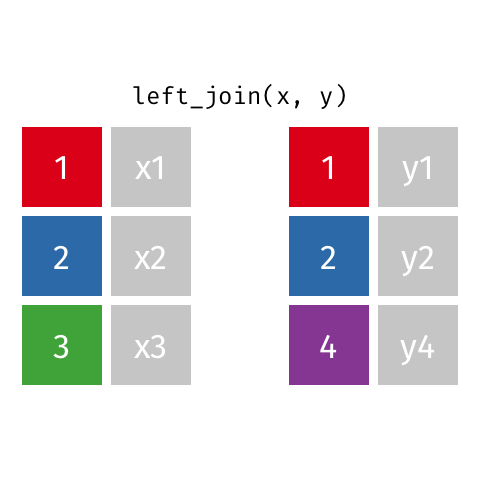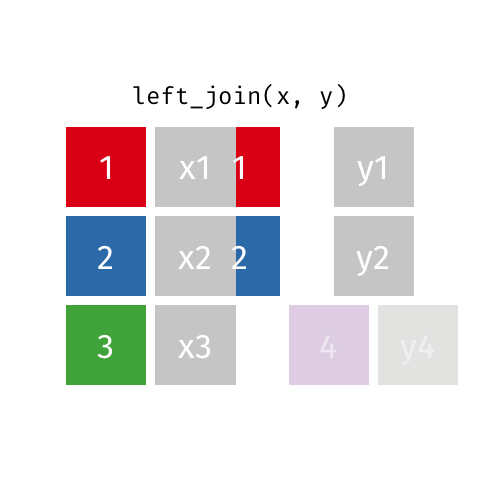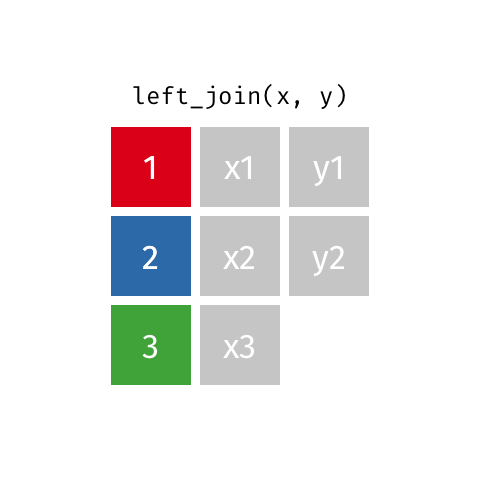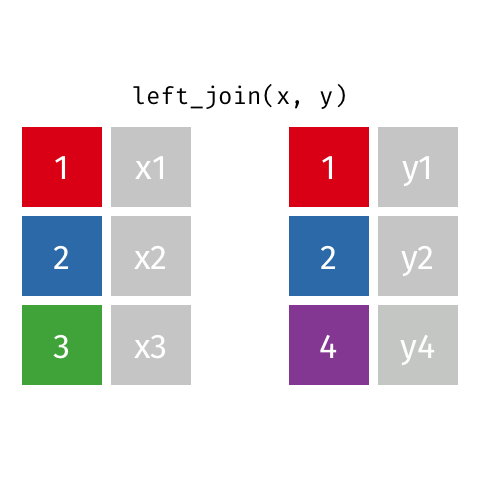
*Animated gif by Garrick Aden-Buie*
`left_join` returns all rows from the first data frame regardless of whether there is a match in the second data frame. Rows with no match are included in the resulting data frame but have `NA` values in the additional columns coming from the second data frame.
Animations to illustrate other types of join are available at <https://github.com/gadenbuie/tidy-animated-verbs>
```{r}
left_join(gene_results, gene_anno)
```
`right_join` is similar but returns all rows from the second data frame that have a match with rows in the first data frame based on the specified column.
```{r}
right_join(gene_results, gene_anno)
```
`inner_join` only returns those rows where matches could be made
```{r}
inner_join(gene_results, gene_anno)
```
------------------------------------------------------------------------
------------------------------------------------------------------------
------------------------------------------------------------------------
# Wrap-up
We have introduced a few of the essential packages from the R tidyverse that can help with data manipulation and visualisation.
 Hopefully you will feel more confident about importing your data into R and producing some useful visualisations. You will probably have questions regarding the analysis of your own data. Some good starting points to get help are listed below.
::: information
- [tidyverse homepage](https://www.tidyverse.org/)
- [R graph gallery](https://www.r-graph-gallery.com/)
:::
To finish the workshop we will look at the analysis of some relevant data that we can import into R and analyse with the tools from the workshop.
## Introducing the COVID-19 data
Data for global COVID-19 cases are available online from CSSE at Johns Hopkins University on their github repository.
::: information
[github](www.github.com) is an excellent way of making your code and analysis available for others to reuse and share. Private repositories with restricted access are also available. Here is a useful beginners guide.
\-[Friendly github intro](https://kirstiejane.github.io/friendly-github-intro/)
:::
R is capable of downloading files to our own machine so we can analyse them. We need to know the URL (for the COVID data we can find this from github, or use the address below) and can specify what to call the file when it is downloaded.
```{r}
download.file("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv",destfile = "raw_data/time_series_covid19_confirmed_global.csv")
```
We can use the `read_csv` function as before to import the data and take a look. We can see the basic structure of the data is one row for each country / region and columns for cases on each day.
```{r}
covid <- read_csv("raw_data/time_series_covid19_confirmed_global.csv")
covid
```
We can potentially join these data to `gapminder`, and it would be beneficial to have one column name in common between both files. We can `rename` the `Country/Region` column of our new data frame to match `gapminder`.
```{r}
covid <- read_csv("raw_data/time_series_covid19_confirmed_global.csv") %>%
rename(country = `Country/Region`)
covid
```
Much of the analysis of this dataset has looked at trends over time (e.g. increasing /decreasing case numbers, comparing trajectories). As we know by now, the `ggplot2` package allows us to map columns (variables) in our dataset to aspects of the plot.
In other words, we would expect to create plots by writing code such as:-
ggplot(covid, aes(x = Date, y =...)) + ...
Unfortunately such plots are not possible with the data in it's current format. Counts for each date are containing in a different column. What we require is a column to indicate the date, and the corresponding count in the next column. Such data arrangements are known as *long data*; whereas we have *wide* data. Fortunately we can convert between the two using the `tidyr` package (also part of tidyverse).
```{r eval=FALSE}
## install tidyr if you don't already have it
install.packages("tidyr")
```
::: information
For more information on *tidy data*, and how to convert between long and wide data, see
<https://r4ds.had.co.nz/tidy-data.html>
:::
```{r}
library(tidyr)
covid <- read_csv("raw_data/time_series_covid19_confirmed_global.csv") %>%
rename(country = `Country/Region`) %>%
pivot_longer(5:last_col(),names_to="Date", values_to="Cases")
covid
```
Another point to note is that the dates are not in an internationally recognised format, which could cause a problem for some visualisations that rely on date order. We can fix by explicitly converting to YYYY-MM-DD format.
<div class="information">
For more ways of dealing with dates in R see the `lubridate` package.
</div>
```{r}
covid <- read_csv("raw_data/time_series_covid19_confirmed_global.csv") %>%
rename(country = `Country/Region`) %>%
pivot_longer(5:last_col(),names_to="Date", values_to="Cases") %>%
mutate(Date=as.Date(Date,"%m/%d/%y"))
covid
```
Another useful modification is to make sure only one row exists for each country. If we look at the data for some countries (e.g. China and UK) there are different entries for provinces and oversees territories.
```{r}
## the count function tabulates the number of observations in a particular column
filter(covid, country == "China") %>%
count(`Province/State`)
```
So we can change the Cases to be the `sum` of all cases for that country on a particular day. We can do this using the `group_by` and `summarise` functions from above
```{r}
covid <- read_csv("raw_data/time_series_covid19_confirmed_global.csv") %>%
rename(country = `Country/Region`) %>%
pivot_longer(5:last_col(),names_to="Date", values_to="Cases") %>%
mutate(Date=as.Date(Date,"%m/%d/%y")) %>%
group_by(country,Date) %>%
summarise(Cases = sum(Cases))
covid
```
### Exercise
::: exercise
What plots and summaries can you make from these data?
- Plotting the number of cases over time for certain countries
- Which country in each continent currently has the highest number of cases?
- Normalise the number of cases for population size (using 2007 population figures as a population estimate)?
- e.g. cases per 100,000
- Which European countries have the highest number of cases per 100,000 population
- e.g. <https://www.statista.com/statistics/1110187/coronavirus-incidence-europe-by-country/>
:::
```{r}
```