-
Notifications
You must be signed in to change notification settings - Fork 31
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Merge pull request #797 from ICB-DCM/release_0.10.11
Release 0.10.11 * Fixed setting initial conditions for preequilibration (#784) * Fixed species->parameter conversion during PEtab import (#782) * Set correct Matlab include directories in CMake (#793) * Extended and updated documentation (#785, #787) * Fix various SBML import issues * Run SBML test suite using github actions instead of travisCI (#789)
- Loading branch information
Showing
49 changed files
with
1,426 additions
and
539 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,24 @@ | ||
| name: SBML semantic test suite | ||
| on: | ||
| push: | ||
| branches: | ||
| - develop | ||
| - master | ||
| - test_action | ||
| pull_request: | ||
| branches: | ||
| - master | ||
|
|
||
| jobs: | ||
| build: | ||
|
|
||
| runs-on: ubuntu-latest | ||
|
|
||
| steps: | ||
| - uses: actions/checkout@v1 | ||
| with: | ||
| fetch-depth: 1 | ||
| - name: apt | ||
| run: sudo apt-get install -y swig3.0 libatlas-base-dev && sudo ln -s /usr/bin/swig3.0 /usr/bin/swig | ||
| - run: AMICI_PARALLEL_COMPILE=2 ./scripts/installAmiciSource.sh | ||
| - run: AMICI_PARALLEL_COMPILE=2 ./scripts/run-SBMLTestsuite.sh |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,29 +1,12 @@ | ||
| # How to contribute | ||
|
|
||
| We are happy about contributions to AMICI in any form (new functionality, documentation, bug reports, ...). | ||
| We are happy about contributions to AMICI in any form, be it new functionality, | ||
| documentation, bug reports, or anything else. | ||
|
|
||
| ## Making code changes | ||
| If you would to contribute to AMICI, a good start is looking for issues tagged | ||
| [`good first issue`](https://github.com/ICB-DCM/AMICI/issues?q=is%3Aissue+is%3Aopen+label%3A%22good+first+issue%22) | ||
| or [`help wanted`](https://github.com/ICB-DCM/AMICI/issues?q=is%3Aissue+is%3Aopen+label%3A%22help+wanted%22). | ||
| For other ideas or questions, just post an issue. | ||
|
|
||
| When making code changes: | ||
|
|
||
| * Check if you agree to release your contribution under the conditions provided in `LICENSE` | ||
| * Start a new branch from `develop` | ||
| * Implement your changes | ||
| * Submit a pull request to the `develop` branch | ||
| * Make sure your code is documented appropriately | ||
| * Run `mtoc/makeDocumentation.m` to check completeness of your documentation | ||
| * Make sure your code is compatible with C++11, `gcc` and `clang` | ||
| * when adding new functionality, please also provide test cases (see `tests/cpputest/`) | ||
| * Write meaningful commit messages | ||
| * Run all tests to ensure nothing was broken | ||
| * Run `tests/cpputest/wrapTestModels.m` followed by CI tests `scripts/buildAll.sh && scripts/run-cpputest.sh` | ||
| * Run `tests/testModels.m` | ||
| * Run `make python-tests` in `build` | ||
| * When all tests are passing and you think your code is ready to merge, request a code review | ||
|
|
||
| ## Adding/Updating tests | ||
|
|
||
| To add new tests add a new corresponding python script (see, e.g., `tests/example_dirac.py`) and add it to and run `tests/generateTestConfigurationForExamples.sh` | ||
| To update test results replace `tests/cpputest/expectedResults.h5` by `tests/cpputest/writeResults.h5.bak` [ONLY DO THIS AFTER TRIPLE CHECKING CORRECTNESS OF RESULTS] | ||
| Before replacing the test results, confirm that only expected datasets have changed, e.g. using | ||
| `h5diff -v -r 1e-8 tests/cpputest/expectedResults.h5 tests/cpputest/writeResults.h5.bak | less` | ||
| For code contributions, please read our | ||
| [developer's guide](documentation/development.md) first. |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,36 +1,133 @@ | ||
| # About AMICI | ||
|
|
||
| AMICI provides a multi-language (Python, C++, Matlab) interface for the | ||
| [SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers | ||
| [CVODES](https://computing.llnl.gov/projects/sundials/cvodes) | ||
| (for ordinary differential equations) and | ||
| [IDAS](https://computing.llnl.gov/projects/sundials/idas) | ||
| (for algebraic differential equations). AMICI allows the user to read | ||
| differential equation models specified as [SBML](http://sbml.org/) | ||
| and automatically compiles such models as `.mex` simulation files | ||
| (Matlab), C++ executables or Python modules. | ||
|
|
||
| AMICI provides a multilanguage (Python, C++, Matlab) interface for the SUNDIALS solvers CVODES (for ordinary differential equations) and IDAS (for algebraic differential equations). AMICI allows the user to read differential equation models specified as SBML and automatically compiles such models as .mex simulation files, C++ executables or python modules. In contrast to the SUNDIALSTB interface, all necessary functions are transformed into native C++ code, which allows for a significantly faster simulation. Beyond forward integration, the compiled simulation file also allows for forward sensitivity analysis, steady state sensitivity analysis and adjoint sensitivity analysis for likelihood based output functions. | ||
| In contrast to the (no longer maintained) | ||
| [sundialsTB](https://computing.llnl.gov/projects/sundials/sundials-software) | ||
| Matlab interface, all necessary functions are transformed into native | ||
| C++ code, which allows for a significantly faster simulation. | ||
|
|
||
| The interface was designed to provide routines for efficient gradient computation in parameter estimation of biochemical reaction models but is also applicable to a wider range of differential equation constrained optimization problems. | ||
| Beyond forward integration, the compiled simulation file also allows for | ||
| forward sensitivity analysis, steady state sensitivity analysis and | ||
| adjoint sensitivity analysis for likelihood based output functions. | ||
|
|
||
| Online documentation is available as [github-pages](http://icb-dcm.github.io/AMICI/). | ||
| The interface was designed to provide routines for efficient gradient | ||
| computation in parameter estimation of biochemical reaction models but | ||
| it is also applicable to a wider range of differential equation | ||
| constrained optimization problems. | ||
|
|
||
| ## Publications | ||
|
|
||
| [](https://zenodo.org/badge/latestdoi/43677177) | ||
|
|
||
| [Fröhlich, F., Kaltenbacher, B., Theis, F. J., & Hasenauer, J. (2017). Scalable Parameter Estimation for Genome-Scale Biochemical Reaction Networks. Plos Computational Biology, 13(1), e1005331. doi: 10.1371/journal.pcbi.1005331](https://doi.org/10.1371/journal.pcbi.1005331) | ||
| ## Features | ||
|
|
||
| [Fröhlich, F., Theis, F. J., Rädler, J. O., & Hasenauer, J. (2017). Parameter estimation for dynamical systems with discrete events and logical operations. Bioinformatics, 33(7), 1049-1056. doi: 10.1093/bioinformatics/btw764](https://doi.org/10.1093/bioinformatics/btw764) | ||
| * SBML import (see details below) | ||
| * Generation of C++ code for model simulation and sensitivity | ||
| computation | ||
| * Access to and high customizability of CVODES and IDAS solver | ||
| * Python, C++, Matlab interface | ||
| * Sensitivity analysis | ||
| * forward | ||
| * steady state | ||
| * adjoint | ||
| * first- and second-order | ||
| * Pre-equilibration and pre-simulation conditions | ||
| * Support for | ||
| [discrete events and logical operations](https://academic.oup.com/bioinformatics/article/33/7/1049/2769435) | ||
|
|
||
| [Full list](documentation/references.md) | ||
|
|
||
| ## Interfaces & workflow | ||
|
|
||
| The AMICI workflow starts with importing a model from either | ||
| [SBML](http://sbml.org/) (Matlab, Python) or a Matlab definition of the | ||
| model (Matlab-only). From this input, all equations for model simulation | ||
| are derived symbolically and C++ code is generated. This code is then | ||
| compiled into a C++ library, a Python module, or a Matab mex file and | ||
| is then used for model simulation. | ||
| compiled into a C++ library, a Python module, or a Matlab `.mex` file and | ||
| is then used for model simulation. | ||
|
|
||
| 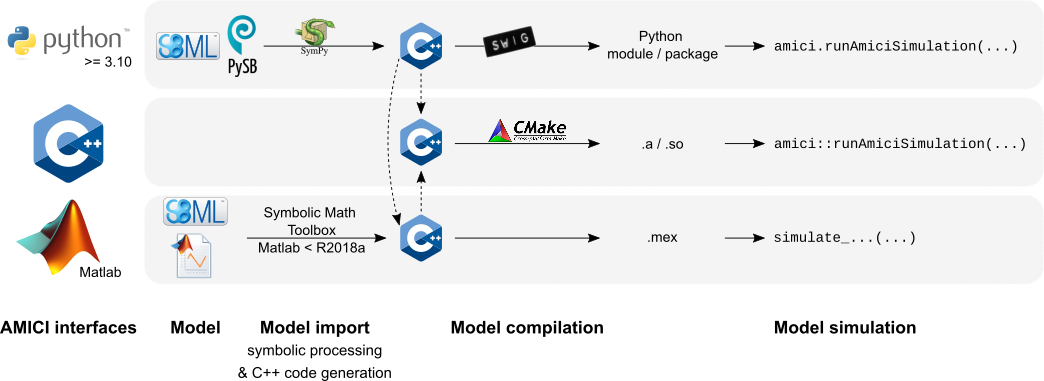 | ||
|
|
||
| ## Getting started | ||
|
|
||
| AMICI installation instructions are provided | ||
| [here](http://icb-dcm.github.io/AMICI/md__i_n_s_t_a_l_l.html). | ||
|
|
||
| To get you started with Python-AMICI the best way might be this | ||
| [Jupyter notebook](https://github.com/ICB-DCM/AMICI/blob/master/python/examples/example_steadystate/ExampleSteadystate.ipynb). | ||
|
|
||
| For Matlab, various examples are available | ||
| [here](https://github.com/ICB-DCM/AMICI/tree/master/matlab/examples). | ||
|
|
||
|
|
||
| Comprehensive documentation on installation and usage of AMICI is available | ||
| online at [http://icb-dcm.github.io/AMICI/](http://icb-dcm.github.io/AMICI/). | ||
|
|
||
| Any contributions to AMICI are welcome, read more contributing | ||
| [here](http://icb-dcm.github.io/AMICI/md__c_o_n_t_r_i_b_u_t_i_n_g.html). | ||
|
|
||
|
|
||
| ### Getting help | ||
|
|
||
| In case of questions or problems with using AMICI, feel free to post an | ||
| [issue](https://github.com/ICB-DCM/AMICI/issues) on Github. We are trying to | ||
| get back to you quickly. | ||
|
|
||
| ## Publications | ||
|
|
||
| **Citeable DOI for the latest AMICI release:** | ||
| [](https://zenodo.org/badge/latestdoi/43677177) | ||
|
|
||
| There is a list of [publications using AMICI](documentation/references.md). | ||
| If you used AMICI in your work, we are happy to include | ||
| your project, please let us know via a Github issue. | ||
|
|
||
| When using AMICI in your project, please cite | ||
| * [Fröhlich, F., Kaltenbacher, B., Theis, F. J., & Hasenauer, J. (2017). Scalable Parameter Estimation for Genome-Scale Biochemical Reaction Networks. Plos Computational Biology, 13(1), e1005331. doi: 10.1371/journal.pcbi.1005331](https://doi.org/10.1371/journal.pcbi.1005331) | ||
| and/or | ||
| * [Fröhlich, F., Theis, F. J., Rädler, J. O., & Hasenauer, J. (2017). Parameter estimation for dynamical systems with discrete events and logical operations. Bioinformatics, 33(7), 1049-1056. doi: 10.1093/bioinformatics/btw764](https://doi.org/10.1093/bioinformatics/btw764) | ||
|
|
||
|
|
||
| ## Status of SBML support in Python-AMICI | ||
|
|
||
| Python-AMICI currently passes 494 out of the 1780 (~28%) test cases from | ||
| the semantic | ||
| [SBML Test Suite](https://github.com/sbmlteam/sbml-test-suite/). | ||
|
|
||
| In additional, we currently plan to add support for the following features | ||
| (see corresponding issues for details and progress): | ||
|
|
||
| - Events (currently Matlab-only) | ||
| - Rate rules | ||
| - Algebraic rules | ||
| - Species assignment rules | ||
| - Compartment assignment rules | ||
| - Models without species | ||
| - Logical operators | ||
|
|
||
| contributions are welcome. | ||
|
|
||
| However, the following features are unlikely to be supported: | ||
|
|
||
| - SBML extensions | ||
| - `factorial()`, `ceil()`, `floor()`, due to incompatibility with | ||
| symbolic sensitivity computations | ||
| - initial assignments for parameters | ||
| - `delay()` due to missing SUNDIALS solver support | ||
|
|
||
|  | ||
|
|
||
| ## Current build status | ||
|
|
||
| [](https://badge.fury.io/py/amici) | ||
| [](https://travis-ci.com/ICB-DCM/AMICI) | ||
| [](https://codecov.io/gh/ICB-DCM/AMICI) | ||
| [](https://www.codacy.com/app/FFroehlich/AMICI?utm_source=github.com&utm_medium=referral&utm_content=ICB-DCM/AMICI&utm_campaign=Badge_Grade) | ||
| <a href="https://badge.fury.io/py/amici" alt="PyPI version"> | ||
| <img src="https://badge.fury.io/py/amici.svg"></a> | ||
| <a href="https://travis-ci.com/ICB-DCM/AMICI" alt="Build Status"> | ||
| <img src="https://travis-ci.com/ICB-DCM/AMICI.svg?branch=master"></a> | ||
| <a href="https://codecov.io/gh/ICB-DCM/AMICI" alt="CodeCov"> | ||
| <img src="https://codecov.io/gh/ICB-DCM/AMICI/branch/master/graph/badge.svg"></a> | ||
| <a href="https://www.codacy.com/app/FFroehlich/AMICI" alt="Codacy"> | ||
| <img src="https://api.codacy.com/project/badge/Grade/945235766e344a7fa36278feab915ff6"></a> |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,79 @@ | ||
| # Continuous integration (CI) and tests | ||
|
|
||
| AMICI uses a continuous integration pipeline running on https://travis-ci.org/. | ||
| This includes the following steps: | ||
|
|
||
| - Checking existence and format of documentation | ||
| - Static code analysis (http://cppcheck.sourceforge.net/) | ||
| - Unit and integration tests | ||
| - Memory leak detection | ||
|
|
||
| More details are provided in the sections below. | ||
|
|
||
| The CI scripts and tests can be found in `tests/` and `scripts/`. Some of the | ||
| tests are integrated with CMake, see `make help` in the build directory. | ||
|
|
||
|
|
||
| ## C++ unit and integration tests | ||
|
|
||
| To run C++ tests, build AMICI with `make` or `scripts/buildAll.sh`, | ||
| then run `scripts/run-cpputest.sh`. | ||
|
|
||
|
|
||
| ## Python unit and integration tests | ||
|
|
||
| To run Python tests, run `../scripts/run-python-tests.sh` from anywhere | ||
| (assumes build directory is `build/`) or run `make python-tests` in your build | ||
| directory. | ||
|
|
||
|
|
||
| ## Matlab tests (not included in CI pipeline) | ||
|
|
||
| To execute the Matlab test suite, run `tests/testModels.m`. | ||
|
|
||
|
|
||
| ## Model simulation integration tests | ||
|
|
||
| Many of our integration tests are model simulations. The simulation results | ||
| obtained from the Python and C++ are compared to results saved in an HDF5 file | ||
| (`tests/cpputest/expectedResults.h5`). | ||
| Settings and data for the test simulations are also specified in this file. | ||
|
|
||
| **Note:** The C++ code for the models is included in the repository under | ||
| `models/`. | ||
| This code is to be updated whenever `amici::Model` changes. | ||
|
|
||
|
|
||
| ### Regenerating C++ code of the test models | ||
|
|
||
| Regeneration of the model code must done whenever `amici::Model` or | ||
| the Matlab model import routines change. | ||
|
|
||
| This is done with | ||
|
|
||
| tests/cpputest/wrapTestModels.m | ||
|
|
||
| **Note:** This is currently only possible from Matlab < R2018a. This should | ||
| change as soon as 1) all second-order sensitivity code is ported to C++/Python, | ||
| 2) a non-SBML import exists for Python and 3) support for events has been added | ||
| for Python. | ||
|
|
||
|
|
||
| ### Regenerating expected results | ||
|
|
||
| To update test results, run `make test` in the build directory, | ||
| replace `tests/cpputest/expectedResults.h5` by | ||
| `tests/cpputest/writeResults.h5.bak` | ||
| [ONLY DO THIS AFTER TRIPLE CHECKING CORRECTNESS OF RESULTS] | ||
| Before replacing the test results, confirm that only expected datasets have | ||
| changed, e.g. using | ||
|
|
||
| h5diff -v -r 1e-8 tests/cpputest/expectedResults.h5 tests/cpputest/writeResults.h5.bak | less | ||
|
|
||
|
|
||
| ## Adding/Updating tests | ||
|
|
||
| To add new tests add a new corresponding python script (see, e.g., | ||
| `./tests/generateTestConfig/example_dirac.py`) and add it to and run | ||
| `tests/generateTestConfigurationForExamples.sh`. | ||
| Then regenerate the expected test results (see above). |
Oops, something went wrong.