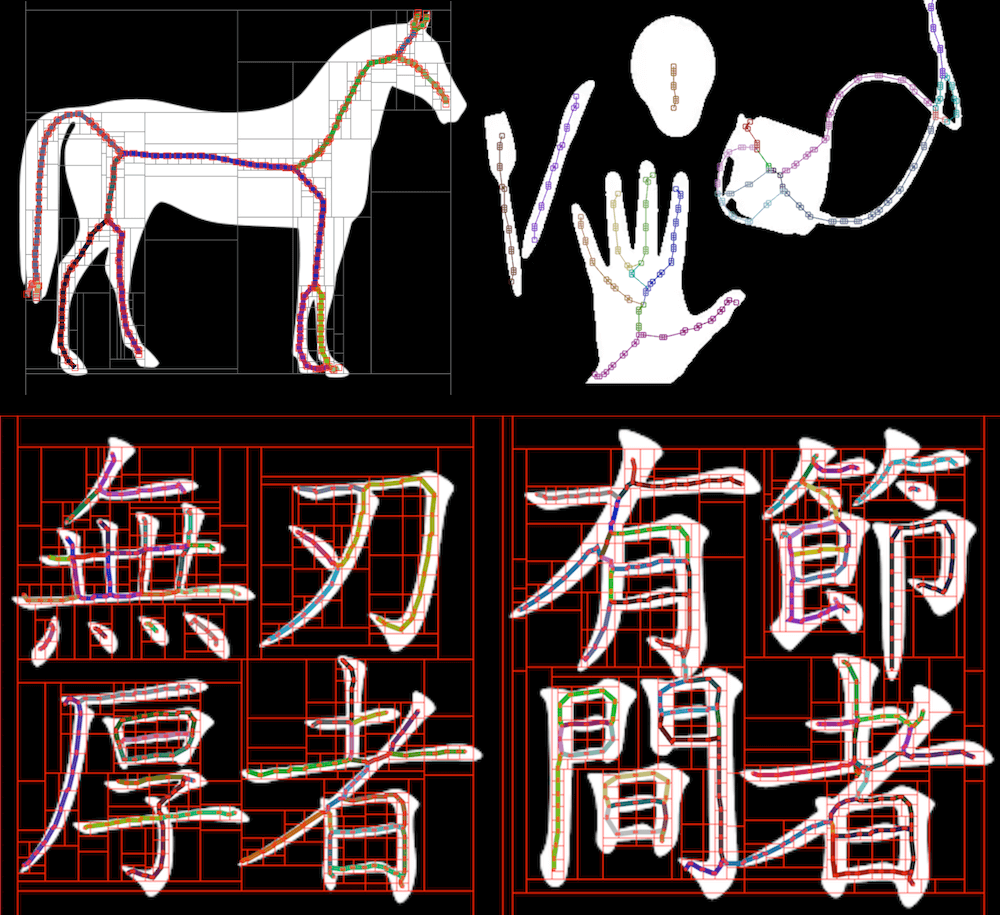
A new algorithm for retrieving topological skeleton as a set of polylines from binary images.
Available in all your favorite languages: C, C++, Java, JavaScript, Python, Go, C#/Unity, Swift, Rust, Julia, WebAssembly, Haxe, Processing, OpenFrameworks.
About the Chinese characters in the test image
Traditionally, skeletonization (thinning) is a morphological operation to reduce a binary image to its topological skeleton, returning a raster image as result. However, sometimes a vector representation (e.g. polylines) is more desirable. Though contour-finding can be used to further trace the results, they usually give enclosing outlines instead of single strokes, and are prone to slight variations in stroke width caused by imperfection in the skeletonization process. In this demo we present a parallelizable divide-and-conquer based algorithm for skeleton tracing, which converts binary images into a set of polylines, i.e. arrays of (x,y) coordinates along the skeleton, in real time.
Define a binary image to be a 2D matrix consisting of 0-pixels(background) and 1-pixels(foreground). The algorithm can be summarized as follows:
- Given a binary image, first skeletonize it with a traditional raster skeletonization algorithm, e.g. Zhang-Suen 1984 is used in this demo. (Without this step, the algoirthm still works to a certian extent, though the quality is generally reduced.)
- If the width and height of the image are both smaller than a small, pre-determined size, go to step 7.
- Raster scan the image to find a row or column of pixels with qualities that best match the following:
- Has the least amount of 1-pixels on itself.
- The 2 submatrices divided by this row or column do not have 1-pixels on their four corners.
- When two or more candidates are found, pick the one that is closer to the center of the image.
- Split the image by this column or row into 2 submatrices (either left and right, or top and bottom depending on whether row or column is selected in the previous step).
- Check if either of the 2 submatrices is empty (i.e. all 0-pixels). For each non-empty submatrix, recursively process it by going to step 2.
- Merge the result from the 2 submatrices, and return the combined set of polylines.
- For each polylines from one submatrix whose either endpoint coincide with the splitting row or column, find another polyline in the other submatrix whose endpoint meets it. If the matrix was split horizontally, then the x-coordinate of the endpoints should differ by exactly 1, and y-coordinate can differ between 0 to about 4 (depending on the steepness of the stroke potrayed), The reverse goes for vertical splitting.
- Recursive bottom. Walk around the 4 edges of this small matrix in either clockwise or ant-clockwise order inspecting the border pixels.
- Initially set a flag to false, and whenever a 1-pixel is encountered whilst the flag is false, set the flag to true, and push the coordinate of the 1-pixel to a stack.
- Whenever a 0-pixel is encountered whilst the flag is true, pop the last coordinate from the stack, and push the midpoint between it and the current coordinate. Then set the flag to false.
- After all border pixels are visited, the stack now holds coordinates for all the "outgoing" (or "incoming") pixels from this small image section. By connecting these coordinates with the center coordinate of the image section, an estimated vectorized representation of the skeleton in this area if formed by these line segments. We further improve the estimate using the following heuristics:
- If there are exactly 2 outgoing pixels. It is likely that the area holds a straight line. We return a single segment connecting these 2 pixels.
- If there are 3 or more outgoing pixels, it is likely that the area holds an intersection, or "crossroad". We do a convolution on the matrix to find the 3x3 submatrix that contains the most 1-pixels. Set the center of all the segments to the center of the 3x3 submatrix and return.
- If there are only 1 outgoing pixels, return the segment that connects it and the center of the image section.
Click on links below to see each implementation's documentation and code.
- C99 (parallelized with pthreads, libpng for reading and X11 for display)
- C++ (thinly wrapped from C version)
- JavaScript (WebAssembly compiled from C++ using emscripten)
- Vanilla JS (Pure JavaScript implementation)
- Pure Python (slow)
- Python using C API (compiled from C using SWIG, compatible with numpy and opencv)
- Java (includes a Processing demo)
- OpenFrameworks addon (friendly wrapper on C++ version)
- C# (demo script for Unity Engine)
- Go (parallelized with goroutines)
- Swift (demo with NSImage and AppKit)
- Rust (simple rust implementation)
- Julia (julia implementation with array views)
The benchmarks below are produced on MacBook Pro Mid 2015 (2.5 GHz Intel Core i7, 16GB 1600 MHz DDR3). The input image is test_images/opencv-thinning-src-img.png (300x149px).
All the times refer to pure or "vanilla" implemenations, not wrappers of C/C++. Wrappers on C/C++ should be comparable to the performance of C plus some overhead. Exception is WebAssembly performance which depends heavily on browser and number of open tabs.
All the times refer to the "tracing" operation only, excluding the raster thinning step (which is not my algorithm (problem), but note that practically, raster thinning often takes longer than the tracing, especially for images with lots of white pixels).
For compiled languages, the highest possible optimization level is selected, unless otherwise specified.
Ordered by fastest to slowest.
| Language | Seconds/1000 Runs | FPS | % C speed | Note |
|---|---|---|---|---|
| C | 0.759748 | 1316 | 100% | -O3 |
| Go | 1.1165248 | 895 | 68% | |
| Rust | 1.39878 | 714 | 54% | -O |
| Java | 1.722 | 580 | 44% | |
| Swift | 1.795619 | 556 | 42% | -O |
| JavaScript | 1.948 | 513 | 38% | Node.js v12.10 |
| C# | 4.266101 | 234 | 17% | Unity 2018.4.8f1 |
| Julia | 4.722 (2.791 amortized) | 211 | 16% | First frame takes 2 sec, the rest 0.002 sec |
| Python | 1015.04818 | 1 | 0% | Python 3.7 |
Developed at Frank-Ratchye STUDIO for Creative Inquiry at Carnegie Mellon University.