-
Notifications
You must be signed in to change notification settings - Fork 9
01 Setting Project Configurations
As many other Snakemake workflow, BGCFlow find information about the input and workflow setting inside the config/ folder. This folder will have a .yaml file with the workflow configurations and metadata, which also points to the samples table containing the input paths.
To load an example configuration, run this wrapper command:
bgcflow initThe above command will create a new file in config/config.yaml.
Note: You should run this command inside the BGCFlow directory or use the arguments
--bgcflow_dir <my BGCFlow folder>
More about the init command:
Usage: bgcflow init [OPTIONS]
Create projects or initiate BGCFlow config from template. Use --project to
create a new BGCFlow project.
Usage: bgcflow init --> check current directory for existing config dir. If
not found, generate from template. bgcflow init --project <TEXT> -->
generate a new BGCFlow project in the config directory.
Options:
--bgcflow_dir TEXT Location of BGCFlow directory. (DEFAULT: Current
working directory)
--project TEXT Initiate a new BGCFlow project. Insert project name:
`bgcflow init --project <TEXT>`
--use_project_pipeline Generate pipeline selection template in PEP file
instead of using Global pipelines. Use with
`--project` option.
--prokka_db TEXT Path to custom reference file. Use with `--project`
option.
--gtdb_tax TEXT Path to custom taxonomy file. Use with `--project`
option.
--samples_csv TEXT Path to samples file. Use with `--project` option.
-h, --help Show this message and exit.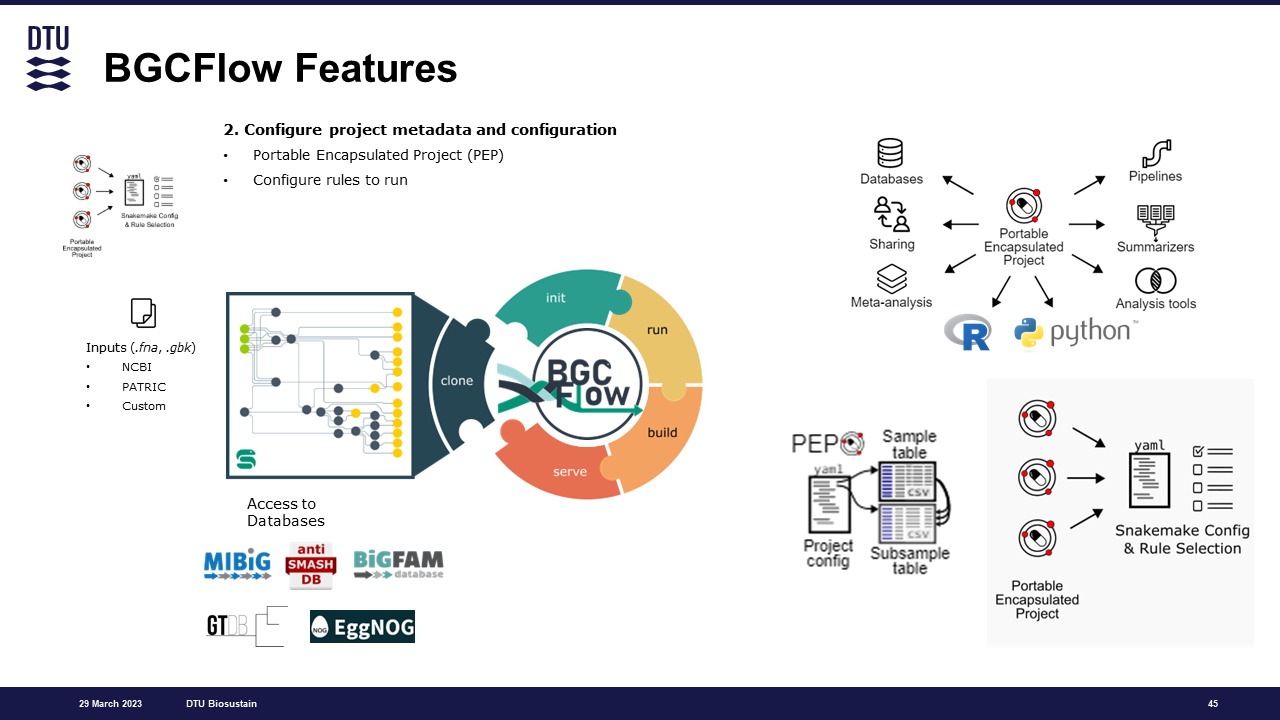
BGCFlow have two different configuration levels, global and project-specific. Both are defined as a .yaml format and structured like below:
config/
├── config.yaml # --> GLOBAL CONFIGURATION FILE
└── project_1
├── project_config.yaml # --> PROJECT CONFIGURATION FILE
└── samples.csvThe global configuration is defined in the config.yaml under the config folder. It's function is to:
- List projects that should be run in the main workflow and subworkflows
- Set up default pipelines/rules that will be run for all projects
- Locate the
resourcespath - Manage other settings that applies to all projects
Configure the workflow according to your needs by editing the files in the config/ folder. An example of the configuration files is provided in the .examples folder.
Projects can be added under the project section of the global config file: config/config.yaml. Each can project can be added as a line containing a path to the project specification configuration files (the PEP file). Each line starts with "-" and the variable pep which points to a PEP config file.
projects:
- pep: .examples/_pep_example/project_config.yamlIn the global config file, you can choose which analysis to run by setting the parameter value in pipelines section to TRUE or FALSE:
pipelines:
bigscape: TRUE
mlst: TRUE
refseq_masher: TRUE
seqfu: TRUE
eggnog: FALSENote that this only applies to the pipelines availaible in the main workflow.
TIPS - Find available rules from the main workflow with
bgcflow_wrapper
$ bgcflow pipelines --bgcflow_dir bgcflow
Printing available rules:
- eggnog
- mash
- fastani
- automlst-wrapper
- roary
- eggnog-roary
- seqfu
- bigslice
- query-bigslice
- checkm
- gtdbtk
- prokka-gbk
- antismash
- arts
- deeptfactor
- deeptfactor-roary
- cblaster-genome
- cblaster-bgc
- bigscapeTIPS - Find out rule description with
bgcflow_wrapper
$ bgcflow pipelines --describe bigscape
Description for bigscape:
- Cluster BGCs using BiG-SCAPE
$ bgcflow pipelines --cite bigscape
Citations for bigscape:
- Navarro-Muñoz, J.C., Selem-Mojica, N., Mullowney, M.W. et al. A computational framework to explore large-scale biosynthetic diversity. [Nat Chem Biol 16, 60–68 (2020)](https://doi.org/10.1038/s41589-019-0400-9)More about the command:
$ bgcflow pipelines --help
Usage: bgcflow pipelines [OPTIONS]
Get description of available pipelines from BGCFlow.
Options:
--bgcflow_dir TEXT Location of BGCFlow directory. (DEFAULT: Current working
directory)
--describe TEXT Get description of a given pipeline.
--cite TEXT Get citation of a given pipeline.
-h, --help Show this message and exit.By default, BGCFlow will download and install necessary softwares and databases in the resources/ folder. The location of each resources can be changed by editing the path in the resource_path section. This is useful, especially if you already have the databases and softwares locally. Instead of creating the resource folder, BGCFlow will generate a symlink to the existing resources.
resources_path:
antismash_db: resources/antismash_db
eggnog_db: resources/eggnog_db
BiG-SCAPE: resources/BiG-SCAPE
bigslice: resources/bigslice
checkm: resources/checkm
gtdbtk: <custom gtdbtk database path>Other configuration is described in the Advanced Configuration page.
As of BGCFlow version >=0.4.0, projects are now configured as a Portable Encapsulated Project (PEP). The project specific configuration is a .yaml file which can be put inside each project folder. It's function is to:
- Define a project id
- Give project description and metadata
- Locate the
samplestable containing a list of the inputs for each project - Add additional information for the workflow run, such as custom
taxonomic assignmentorcustom reference gene annotation - Define pipelines/rules to run for a particular project. This will override and ignore the pipelines/rules defined in the global configuration.
See project_config.yaml for an example of a PEP formatted project.
Each project will requires a name and description. An example project PEP configuration will look like this:
name: Lactobacillus_delbrueckii
pep_version: 2.1.0
description: "Lactobacillus delbrueckii 27 01 2023"
sample_table: samples.csv
#### RULE CONFIGURATION ####
# rules: set value to TRUE if you want to run the analysis or FALSE if you don't
rules:
seqfu: TRUE
mash: TRUE
fastani: TRUE
checkm: FALSEThe name will be used as the project id, and should be unique for each project. The description should be given to provide context about the project, sample size, date of experiment, etc. The variable pep_version will tell BGCFlow which version of PEP is being used. Additional configuration is described in the Advanced Configuration section.
The variable sample_table (PEP) or samples denote the location of your .csv file which specifies the genomes to analyze. Note that you can name the file anything as long as you define it in the config.yaml.
Example: samples.csv
| genome_id | source | organism | genus | species | strain | closest_placement_reference |
|---|---|---|---|---|---|---|
| GCF_000359525.1 | ncbi | J1074 | ||||
| 1223307.4 | patric | Streptomyces sp. PVA 94-07 | Streptomyces | sp. | PVA 94-07 | GCF_000495755.1 |
| P8-2B-3.1 | custom | Streptomyces sp. P8-2B-3 | Streptomyces | sp. | P8-2B-3 |
Columns description:
-
genome_id[required]: The genome accession ids (with genome version forncbiandpatricgenomes). Forcustomfasta file provided by users, it should refer to the fasta file names stored in thedata/raw/fasta/directory with.fnaextension. Example: genome id P8-2B-3.1 refers to the filedata/raw/fasta/P8-2B-3.1.fna. -
source[required]: Source of the genome to be analyzed choose one of the following:custom,ncbi,patric. Where:-
custom: for user-provided genomes (.fna) in thedata/raw/fastadirectory with genome ids as filenames -
ncbi: for list of public genome accession IDs that will be downloaded from the NCBI refseq (GCF...) or genbank (GCA...) database -
patric: for list of public genome accession IDs that will be downloaded from the PATRIC database
-
-
organism[optional]: name of the organism that is the same as in the fasta header -
genus[optional] : genus of the organism. Ideally identified with GTDBtk. -
species[optional]: species epithet (the second word in a species name) of the organism. Ideally identified with GTDBtk. -
strain[optional] : strain id of the organism -
closest_placement_reference[optional]: if known, the closest NCBI genome to the organism. Ideally identified with GTDBtk.
Further formatting rules are defined in the workflow/schemas/ folder.
In each projects, you can choose which analysis to run by setting the parameter value in the project_config.yaml to TRUE or FALSE:
rules:
bigscape: TRUE
mlst: TRUE
refseq_masher: TRUE
seqfu: TRUE
eggnog: FALSEThis will ignore the pipelines configuration set in the global configuration.