Arun Sethuraman, PhD San Diego State University [email protected] Twitter: @arunsethuraman
- Review performing tests of Hardy-Weinberg Equilibrium, Linkage Disequilibrium, and Differentiation
- Learn how to correct for multiple testing
- Compare results and inferences between different correction techniques, as against uncorrected tests
- vcftools - https://vcftools.github.io/index.html
- Base R or RStudio
- Input files - accessible on the Box page under Lecture Materials > Arun Sethuraman > Hands-on Files
We will be using the data from Bataillon et al. 2015 (Bataillon, T., Duan, J., Hvilsom, C., Jin, X., Li, Y., Skov, L., Glemin, S., Munch, K., Jiang, T., Qian, Y. and Hobolth, A., 2015. Inference of purifying and positive selection in three subspecies of chimpanzees (Pan troglodytes) from exome sequencing. Genome biology and evolution, 7(4), pp.1122-1132.) for this exercise. You can download the VCF file here: http://datadryad.org/resource/doi:10.5061/dryad.56m2g or the SNP file is available as "chimps.vcf" in the Hands-on Files folder.
Back story - this study analyzes whole exome data from three subspecies of chimpanzees - Pan troglodytes troglodytes (Central), P.t.verus (Western), and P.t.schweinfurthii (Eastern) which have very distinctive geographical ranges separated by large river systems in Africa, and have not been observed to hybridize in the wild with each other, or with Pan paniscus (Bonobos, but genomic evidence states otherwise!). Nonetheless, chimpanzee numbers are dwindling in the wild, with estimates of effective population sizes between 22,000 - 27,000 (Fischer et al., 2004, Sethuraman and Hey 2016, Won and Hey 2005).
Further references:
- Won, Y.J. and Hey, J., 2005. Divergence population genetics of chimpanzees. Molecular biology and evolution, 22(2), pp.297-307.
- Sethuraman, A. and Hey, J., 2016. IM a2p–parallel MCMC and inference of ancient demography under the Isolation with migration (IM) model. Molecular ecology resources, 16(1), pp.206-215.
- Kuhlwilm, M., Han, S., Sousa, V.C., Excoffier, L. and Marques-Bonet, T., 2019. Ancient admixture from an extinct ape lineage into bonobos. Nature ecology & evolution, 3(6), pp.957-965.
- Fischer, A., Wiebe, V., Pääbo, S. and Przeworski, M., 2004. Evidence for a complex demographic history of chimpanzees. Molecular biology and evolution, 21(5), pp.799-808.
-
Login to the congen server (handson.congen1.com)
-
Create a new directory within your user directory, and place the SNP VCF file in this folder.
cd /home/username/
mkdir chimps
cd chimps
cp /home/Instructor_Lecture_Materials_and_Hands-on/Arun_Sethuraman/2023/copy_um_box/Hands-on\ Files/* .- Compute heterozygosity across the entire file
vcftools --vcf chimps.vcf --het --out allchroms_het
R #this will open R - or alternately just switch to your Rstudio window on the congen server- Visualize this in R, to observe some basic statistics about the individual chimps.What all statistics were obtained? You’ll notice that the O.HOM. column contains the number of observed homozygote sites, and the E.HOM. contains the number of expected homozygote sites (under HWE), N.SITES is the total number of loci that were analyzed. The first column (INDV) contains the names of all individuals, while the last column, F contains the inbreeding coefficient. What do you observe? Who is most inbred? Who is least inbred?
#In your Rstudio console, click on Session > Set Working Directory > To Files Pane Location (or your chimps directory)
allchroms_het<-read.table("allchroms_het.het",header=TRUE)
allchroms_het
summary(allchroms_het)
plot(allchroms_het[,5],xlab="Individuals",ylab="Genome-wide Heterozygosity")
#q()- Testing for Hardy-Weinberg Equilibrium, computing Diversities, Tajima's D across chromosomes in 10k windows, visualization
#Switch to your Terminal window to run these other commands
vcftools --vcf chimps.vcf --hardy --out allchroms_hwe
vcftools --vcf chimps.vcf --TajimaD 10000 --out allchroms_tajimad
vcftools --vcf chimps.vcf --window-pi 10000 --window-pi-step 10000 --out allchromspi_10kwindow#Switch to the Rstudio console
allchroms_hwe<-read.table("allchroms_hwe.hwe",header=TRUE)
tajimad<-read.table("allchroms_tajimad.Tajima.D",header=TRUE)
tajimad_nomissing<-na.omit(tajimad)
hist(tajimad_nomissing$TajimaD,main="Tajima's D distribution",xlab="Tajima's D",ylab="Frequency")
#Alternately, if we plot it by chromosome
#Plot of Tajima's D
plot(tajimad_nomissing$CHROM,tajimad_nomissing$TajimaD,xlab="Chromosome",ylab="Tajima's D")
#Box Plot of log p-values for sites in HWE
boxplot(log(P_HWE,10)~CHR,data=allchroms_hwe,xlab="Chromosome",ylab="log10 P-value")
These should produce rather ugly plots like the ones below:
So let's make these prettier, and visualize as a Manhattan Plot instead - this requires the qqman package in R. Note that qqman and its dependencies (e.g. calibrate) are available for R.3.5.0 and above - so please note that the Manhattan plots will only work with these versions and above).
#install.packages("qqman")
library(qqman)
manhattan(allchroms_hwe,chr="CHR",bp="POS",p="P_HWE",snp="POS",logp=TRUE,ylab="log p-values",ylim=c(0,10.0))
manhattan(tajimad_nomissing,chr="CHROM",bp="BIN_START",p="TajimaD",snp="N_SNPS",logp=TRUE,ylab="Tajima’s D",ylim=c(-1.0,3.0))Voila! Much prettier(?) What do you notice about these?
Now we'll do the same thing, but with the diversity estimates.
diversity<-read.table("allchromspi_10kwindow.windowed.pi",header=TRUE)
manhattan(diversity,chr="CHROM",bp="BIN_START",p="PI",snp="N_VARIANTS",logp=FALSE,ylab="Pi",ylim=c(0,0.001))Now compare the Tajima's D and Diversity plots. Do you notice anything "cool"?
Now let's try and compute some differentiation statistics based on these data. The three files that are provided (schweinfurthii.txt, troglodytes.txt, and verus.txt) contain the names of each of the individuals from each separate population. Make sure that these files are present in the same folder (chimps) prior to running these commands.
#Move to your terminal window
vcftools --vcf chimps.vcf --weir-fst-pop schweinfurthii.txt --weir-fst-pop troglodytes.txt --weir-fst-pop verus.txt --out allthreepopsfst
#Move onto Rstudio consoleThereon, let's plot and analyze these in R:
fst<-read.table("allthreepopsfst.weir.fst",header=TRUE)
manhattan(fst,chr="CHROM",bp="POS",p="WEIR_AND_COCKERHAM_FST",snp="POS",logp=FALSE,ylab="Fst",ylim=c(0.0,1.1))Now compare this Manhattan plot against the Tajima's D and Diversity plots. Do you notice any patterns?
However, the problem with vcftools is that it doesn't compute p-values for estimates of Tajima's D, diversity, or Fst. So there are other packages that might be used for the same purpose - e.g. hierfstat, ARLEQUIN
- Let's use the same P-values from the tests of HWE - how do we correct for multiple testing in base R?
Turns out, base R has some very useful functions to do this - we will just add new columns to our HWE results, to compare them against each other.
#In Rstudio console
allchroms_hwe$bonferroni=p.adjust(allchroms_hwe$P_HWE,"bonferroni")
allchroms_hwe$holm=p.adjust(allchroms_hwe$P_HWE,"holm")
allchroms_hwe$bh=p.adjust(allchroms_hwe$P_HWE,"BH")
allchroms_hwe$fdr=p.adjust(allchroms_hwe$P_HWE,"fdr")
#Now you can determine which ones are statistically significant at the alpha (FPR) of 0.05
bonferroni<-which(allchroms_hwe$bonferroni < 0.05)
holm<-which(allchroms_hwe$holm < 0.05)
bh<-which(allchroms_hwe$bh < 0.05)
fdr<-which(allchroms_hwe$fdr < 0.05)
x<-data.frame(allchroms_hwe$CHR[bonferroni],allchroms_hwe$POS[bonferroni])
write.table(x,"exclude.txt",row.names=FALSE,quote=FALSE,col.names=FALSE)
#Some examples
head(allchroms_hwe[holm,])
tail(allchroms_hwe[bh,])What do you notice about these loci?
Some thought exercises:
-
Based on your tests of Tajima’s D, do you identify particular loci that have highly positive, and highly negative values across the genome? Make a list of these sites, as well as their coordinates. HINT - you can do this in R by summarizing the Tajima’s D values (using the summary() function), then using the which() function to find out which values are either greater than or less than a “cutoff” value.
-
Repeat the analyses above for identifying so called “outlier” loci/windows that have extremely high levels of diversity or extremely low levels of diversity, and extremely high levels of differentiation, and extremely low levels of differentiation. Are there any overlaps between these lists?
-
Interpret your results - why would you have overlapping regions between these lists at all? Explain.
-
Now do something fun - go to the Ensembl Chimpanzee genome database: https://uswest.ensembl.org/Pan_troglodytes/Info/Index
Now search for some of these loci that you’ve identified as outliers. E.g. The highest Tajima’s D value happens to be 2.97, which is at a window on Chromosome 11, between 56110000 and 56120000. So searching for 11:5611000-5621000 shows up several genes in and around that region. What are some genes that you notice? What would a high degree of positive Tajima’s D at this locus mean?
Now redo this analysis with some other Tajima’s D outliers. What genes do you identify? What do you hypothesize is happening at this locus across chimpanzees?
-
Make sure to have downloaded the data (Exercise2.txt) from the Box page: Lecture Materials > Arun Sethuraman > Hands-on Files > Exercise2.txt
-
In Unix shell, you can "explore" this data by using head/less, or other commands - for instance, here you'll notice that the file contains count information for each genotype - the first column contains the name of the SNP, while the rest of the columns contain genotype counts of A/A, A/B, B/B respectively, where A and B are the two alleles at each locus. For our purpose, we will use the "HardyWeinberg" package in R to test, correct, and obtain "outliers".
-
Now in your Rstudio console:
#install.packages("HardyWeinberg")
library(HardyWeinberg)
x<-read.table("Exercise2.txt")
#note that this is of dimension 5933 x 4 - you can find this by doing dim(x)
pvals<-c() #Empty array to store p-values from HWE tests
i<-1
while (i < 5933) {
pvals[i]<-HWChisq(c(x[i,2],x[i,3],x[i,4]))$pval
i=i+1
}
summary(pvals)- Determine how many loci are out of HWE at an alpha of 0.05, then correct the p-values and then determine the same numbers
length(which(pvals<0.05))
# Turns out there are 1477 loci that are out of HWE out of the 5932, if we don't correct for FPR
# Now let's try a bunch of corrections, to see if we can whittle this down to correct for FPR
bonferroni<-p.adjust(pvals,"bonferroni")
bh<-p.adjust(pvals,"BH")
holm<-p.adjust(pvals,"holm")
#Note that from the manuscript, rs62625034 is the deletion that is determined as the delta32 mutation - so you can search for this particular variant in the dataset, see what you obtain as a p-value prior to correction, and after correction
z<-which(x$V1=="rs62625034")
pvals[z]
which(pvals < pvals[z])What do you notice? Do we recapitulate the results from Wei and Nielsen?
Thought Exercise:
Why or why don't we recapitulate the results from Wei and Nielsen? Can you think of some other ways for us to make this more efficient?
This tutorial has been adapted from the OutFLANK vignette that can be accessed here: https://htmlpreview.github.io/?https://github.com/whitlock/OutFLANK/blob/master/inst/doc/OutFLANKAnalysis.html
First things first, we need to filter our dataset (chimps.vcf) for excessive degrees of missingness, and deviations from HWE prior to running OutFLANK analyses. So to do this, go to your Terminal.
vcftools --vcf chimps.vcf --hwe 0.05 --max-missing 0.25 --recodeThis should create a new filtered VCF file called "out.recode.vcf" in your folder. What do you notice about the number of SNP's in this new file?
#To install OutFLANK:
# devtools::install_github("whitlock/OutFLANK")
library(OutFLANK)
library(vcfR) #should already be installed
#Read the chimps.vcf file, convert it into OutFLANK format
chimps<-read.vcfR("out.recode.vcf")
geno <- extract.gt(chimps) # Character matrix containing the genotypes
position <- getPOS(chimps) # Positions in bp
chromosome <- getCHROM(chimps) # Chromosome information
#Let's specify the population ID's as 1, 2 and 3 for P.t.schweinfurthii, P.t.troglodytes, P.t.verus respectively
pop <- c("1","1","1","1","1","1","1","1","1","1","1","2","2","2","2","2","2","2","2","2","2","2","2","3","3","3","3","3","3","3","3")
G <- matrix(NA, nrow = nrow(geno), ncol = ncol(geno))
G[geno %in% c("0/0", "0|0")] <- 0
G[geno %in% c("0/1", "1/0", "1|0", "0|1")] <- 1
G[geno %in% c("1/1", "1|1")] <- 2
G[is.na(G)]<-9 #important! Missing data calls need to be coded as 9 in OutFLANK
table(as.vector(G))
# Now G should be in OutFLANK format
# Calculate Fst
my_fst<-MakeDiploidFSTMat(t(G), locusNames = position, popNames = pop)
# You can view this
head(my_fst)
#Now run OutFLANK
out_trim<-OutFLANK(my_fst,NumberOfSamples=3,qthreshold=0.05,Hmin=0.1)
head(out_trim$results)
#There are several pruning steps, sanity checks that I am skipping here for time - please refer to the original vignette for more details!
#OutFLANK automates p-value correction for multiple testing using the FDR method, and threshold - please see Whitlock and Lotterhos 2015
P1 <- pOutlierFinderChiSqNoCorr(my_fst, Fstbar = out_trim$FSTNoCorrbar, dfInferred = out_trim$dfInferred, qthreshold = 0.1, Hmin=0.1)
head(P1)
summary(P1$pvalues)Now let's go ahead and plot these:
my_out <- P1$OutlierFlag==TRUE
plot(P1$He, P1$FST, pch=19, col=rgb(0,0,0,0.1))
points(P1$He[my_out], P1$FST[my_out], col="blue")
plot(P1$LocusName[P1$He>0.1], P1$FST[P1$He>0.1],
xlab="Position", ylab="FST", col=rgb(0,0,0,0.2))
points(P1$LocusName[my_out], P1$FST[my_out], col="magenta", pch=20)
Thought Exercise:
What do you determine? Do you find any outlier loci? If you were interpreting these plainly based on p-values, would you identify outliers? What if you had done the same analyses, but with a different q threshold? If you were only using the Tajima's D/Fst/Diversity outliers, how would your inference change?
Arun Sethuraman, PhD San Diego State University [email protected] Twitter: @arunsethuraman
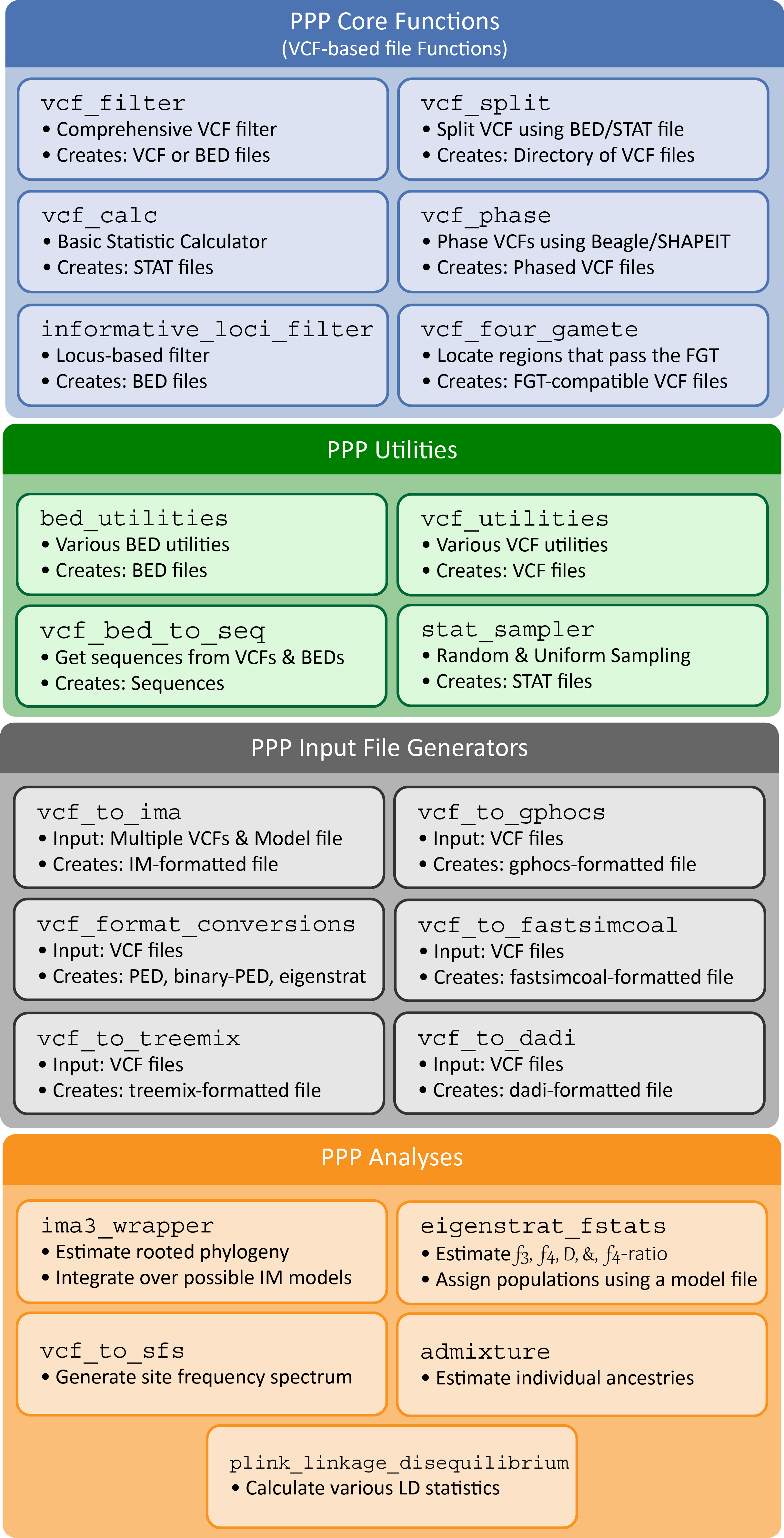
Let's be real - it is becoming increasingly difficult to create reproducible workflows in population genomics. We developed the Popgen Pipeline Platform (PPP) - https://ppp.readthedocs.io/en/latest/index.html with the goal of reducing the computational expertise required for conducting population genomic analyses. The PPP was designed as a collection of scripts that facilitate common population genomic workflows in a consistent and standardized environment. Functions were developed to encompass entire workflows, including: input preparation, file format conversion, various population genomic analyses, and output generation. By facilitating entire workflows, the PPP offers several benefits to prospective end users - it reduces the need of redundant in-house software and scripts that would require development time and may be error-prone, or incorrect, depending on the expertise of the investigator. The platform has also been developed with reproducibility and extensibility of analyses in mind. Better yet, coming 2023 - we have a new Galaxy Project (www.usegalaxy.org) implementation of PPP, which will make your population genomic analyses that much easier with an accessible web-browser based GUI.
We will utilize the same dataset from our first set of exercises to perform a series of analyses using PPP. Specifically, the script below will (1) create different model files (containing population and individual designations), (2) filter the provided VCF file based on a variety of conditions, (3) calculate summary statistics, (4) phase, (5) convert the dataset into a format readable by TreeMix (https://bitbucket.org/nygcresearch/treemix/wiki/Home).
#Let's do some housekeeping of the VCF file first - bgzip + tabix indexing provides a convenient base for all further analyses
bgzip -c chimps.vcf > chimps.vcf.gz
tabix -p vcf chimps.vcf.gz
#Creating Model Files
#Create a model file called 2pop.model only containing the P.t.verus and P.t.troglodytes individuals
model_creator.py --model 2pop --model-pop-file 2pop 2pops.txt --pop-ind-file Verus verus.txt --pop-ind-file Troglodytes troglodytes.txt --out 2pop.model
#Create a model file called 3pop.model all three subspecies
model_creator.py --model 3pop --model-pop-file 3pop 3pops.txt --pop-ind-file Verus verus.txt --pop-ind-file Troglodytes troglodytes.txt --pop-ind-file Schweinfurthii schweinfurthii.txt --out 3pop.model
#Filtering the VCF file to output only bi-allelic sites, and only P.t.verus and P.t.troglodytes individuals
vcf_filter.py --vcf chimps.vcf.gz --filter-only-biallelic --filter-include-indv-file verus.txt --filter-include-indv-file troglodytes.txt --out verustrog.vcf.gz --out-format vcf.gz
#Perform a series of calculations of summary statistics a) Tajima's D in windows of 10kbps, b) per site estimates of Weir and Cockerham's pairwise Fst values between each pair of subspecies
vcf_calc.py --vcf verustrog.vcf.gz --calc-statistic TajimaD --statistic-window-size 10000 --out verustrogtajima
vcf_calc.py --vcf chimps.vcf --model-file 3pop.model --model 3pop --calc-statistic weir-fst --out 3popsfst
#Use SHAPEIT to phase the unphased VCF file - https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html
vcf_phase.py --vcf chimps.vcf --phase-algorithm shapeit
#Convert the VCF file into a format readable by TREEMIX
vcf_to_treemix.py --vcf chimps.vcf.gz --model-file 3pop.model --modelname 3pop --out chimpstreemix
Time to get creative - write a PPP pipeline to perform a set of analyses that you envision would work with your own data (assuming that you are all working with VCF files here). Let's chat!
ConGen2023 - Part 3: Using IMa3 for estimation of demographic history under the Isolation with Migration Model
The Isolation with Migration (IM) class of models (Nielsen and Wakeley 2001) have been utilized extensively since their inception as a framework for inferring evolutionary history of species and populations.

The IM model is ideal for scenarios where you sample and genotype/sequence individuals from: (a) two or more species that are hypothesized to be speciating allopatrically, with continued gene flow (b) two or more populations that are structured/admixed (c) hybrid species (d) species with secondary contact (e) species/populations with hybridization/admixture with unsampled ghost populations Alternately, if you’re a data-up person, IMa3 (and MigSelect, IMa2p) are ideal for analyses of multi-locus genetic/genomic/haplotypic data generated using (a) whole genomic sequencing, (b) Sanger sequencing of individual loci, both nuclear and organellar, (c) RADseq (and variants of it), (d) microsatellite genotyping, to name a few. Importantly, IMa3 (and the IM class of programs) generate evolutionary history, estimated as effective population sizes (or scaled as population mutation rates), divergence times, and migration rates by sampling genealogies (for more details on the broader class of genealogy samplers, see Kuhner 2009). Other genealogy samplers include LAMARC (Kuhner 2006), MIGRATE-n (Beerli and Felsenstein 2001), MIST (Chung and Hey 2017), MDIV (Nielsen and Wakeley 2001), IM (Hey and Nielsen 2004), IMa2 (Hey and Nielsen 2007, Hey 2009). They all broadly utilize a Bayesian (Metropolis-Coupled) Markov Chain Monte Carlo (MCMCMC) algorithm to sample genealogies guided by an evolutionary tree to estimate/approximate the posterior density distribution of evolutionary parameters. They all harken back to the model of Felsenstein 1981:

Consider an alternative expression, that also integrates over G , but that directly yields a posterior probability of Θ

This is an expectation of P(Θ |G) and can be approximated given a sample of genealogies drawn at random from the posterior distribution of G, P(G | X)


This step does not depend on the data, X. All the information in the date is contained in the sample drawn from P(G|X) Yields an analytic function, different from Nielsen & Wakeley (2001) approach which estimates the probability from the residence times in the MCMC Using MCMC to generate samples of genealogies drawn from P(G|X)

• It is not difficult to calculate P(X|G). • But how to calculate the prior probability of G, P(G) ? • In fact this can be calculated analytically for the main demographic components of Θ, assuming a uniform prior for Θ

In short, the sequence of operations performed are: • Run a Markov chain over G and generate random samples from P(G | X) • For each G drawn from this distribution, save P(G) and all necessary information for calculating P(G|Θ). • Build a function that approximates the posterior density of Θ

• This is an analytic function, and can be evaluated for any value of Θ • The function can be differentiated and searched for maxima.
However, these computations and sampling along Metropolis Coupled Markov Chains can be potentially slow and difficult to reach “convergence” or “stationarity”, especially with a large number of (a) loci, (b) individuals, (c) sampled populations. An excellent and straightforward strategy to leverage the power of parallel processing and modern-day high performance computing (HPC) facilities is to distribute these Metropolis Coupled Markov Chains among multiple processors, or “threads”. The most important part of this strategy is to ensure that the sampling process is synchronized across chains. IMa3’s parallel algorithm strategically handles these scenarios of computational difficulties with parallelization, details of which are described in Sethuraman and Hey 2016.


What’s cool about IMa2p and IMa3? Fast, faster, faster…

My goals in this workshop are manifold: (a) help you understand the fundamental algorithm, and parameters estimated by IMa3 (b) describe installation, and computational considerations while working with IMa3 (c) describe all command line options in IMa3, and their individual utilities, by working through some previously simulated examples (d) understand the output files produced by IMa3, and how to interpret them, as well as build your own plots of density distributions in R (e) work through a real life “work-flow”, starting with whole-genomic data, downloaded as a VCF file from The Great Ape Genome Project to estimate the evolutionary history of two species – Pan troglodytes troglodytes, and Pan troglodytes verus. (f) provide suggestions for how you should set up IMa3 runs with your own data (g) briefly introduce IMGui (Knoblauch et al. 2017), and IMfig programs (h) and if we have more time, work through an example of simulating your own genomic data with ms (Hudson 2002) under an IM model, and running IMa3 analyses on these simulated data.
You’ve been provided with an IMa3 formatted input file “wonhey.u” – let’s first look at this file, and understand how this was constructed.
-i specifies the input file
-o specifies the name for the output file(s)
-s random number seed
-r25 specifies some run parameters
-b specifies the length of the “burn-in” period
-l specifies the length of the “run” after “burn-in”
-q specifies an upper limit on theta(s)
-m specifies an upper limit on migration rate(s)
-t specifies an upper limit on the divergence time(s)
-hfg -ha0.99 -hb0.3 - specifies how to run the method
To run IMa3, type:
#Copy the IMa3 input file to your folder
cp /home/Instructor_Lecture_Materials_and_Hands-on/Arun_Sethuraman/2023/copy_um_box/Hands-on\ Files/wonhey.u .
IMa3 -i wonhey.u -o example1.out -b100 -l100 -q10 -m10 -t10 -s124 -r25 -hfg -ha0.99 -hb0.3 -hn10Let’s look at the output files produced (also see sample output file provided for a long run), and understand how to interpret these results.
Let’s then discuss some possible models that can be tested using the LLR test of Nielsen and Wakeley 2001 (in the L mode of IMa3).