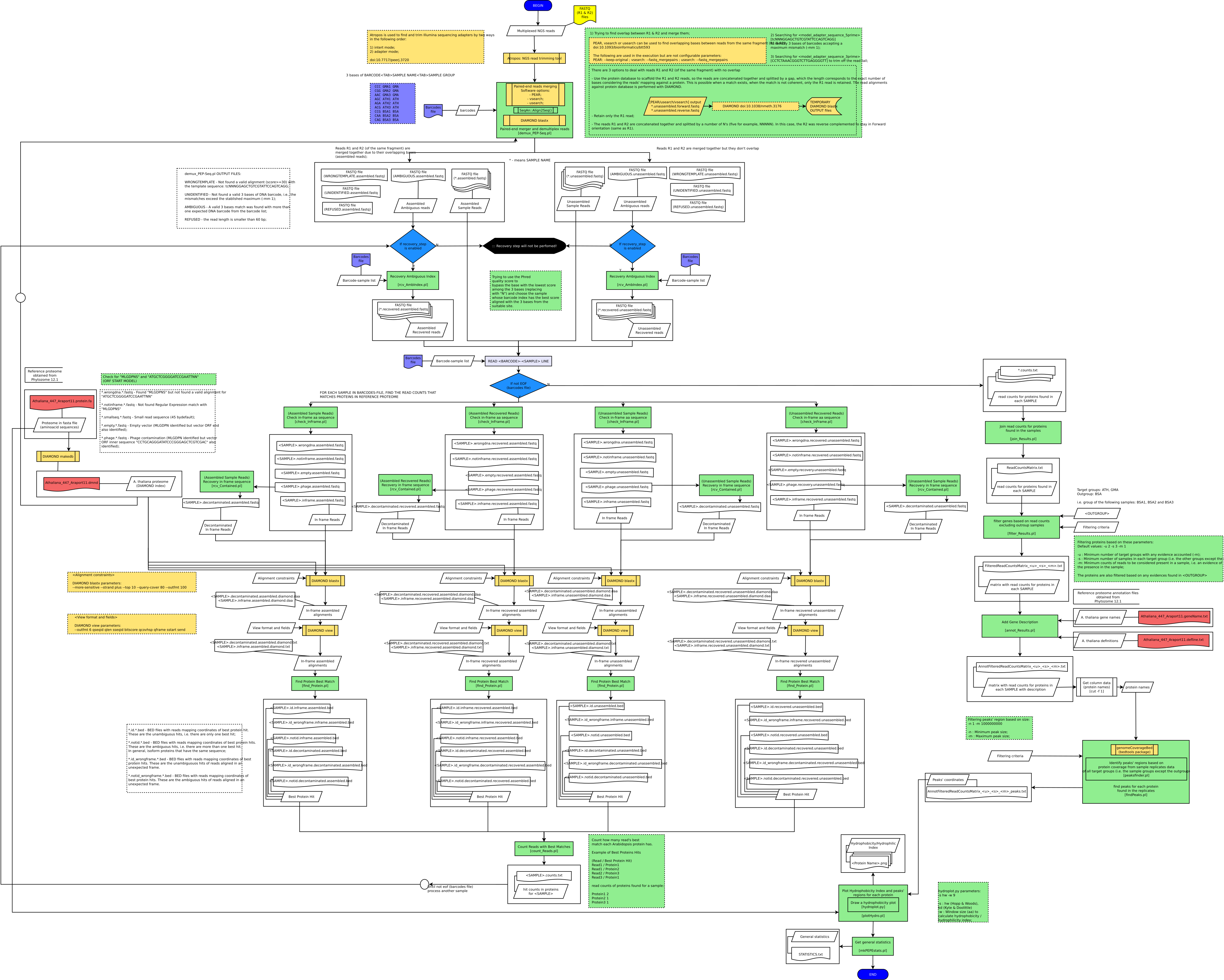
Tools and libraries
DIAMOND - alignment tool for short DNA sequencing reads [v.0.9.22] [https://ab.inf.uni-tuebingen.de/software/diamond]
PEAR - Paired-End reAd mergeR [v.0.9.8] [https://sco.h-its.org/exelixis/web/software/pear/ ]
SeqAn - The Library for Sequence Analysis [v1.4.2] [https://github.com/seqan/seqan]
Perl libraries
Term::ProgressBar [v.2.22] [https://metacpan.org/release/Term-ProgressBar]
Log::Log4perl [v.1.44] [https://metacpan.org/release/Log-Log4perl]
Bio::Root::Version [v.1.006924] [https://metacpan.org/release/BioPerl]
Test::More [v.1.001014] [https://metacpan.org/release/Test-Simple]
Term::ProgressBar [v.2.17] [https://metacpan.org/release/Term-ProgressBar]
FileHandle [v.2.02] [https://metacpan.org/release/perl]
Getopt::Long [v.2.45] [https://metacpan.org/release/Getopt-Long]
File::Basename [v.2.85] [https://metacpan.org/release/perl]
File::Temp [v.0.2304] [https://metacpan.org/release/File-Temp]
FindBin [v.1.51] [https://metacpan.org/release/perl]
XSLoader [v.0.20] [https://metacpan.org/release/XSLoader]
Reference files from Phytozome [https://phytozome.jgi.doe.gov/]:
Files for **Arabidopsis thaliana**:
Athaliana_447_Araport11.protein.fa
Athaliana_447_Araport11.geneName.txt
Athaliana_447_Araport11.defline.txt
Athaliana_447_Araport11.annotation_info.txt
Paired-End fastq input files (R1 & R2, *i.e.* separately):
Before you run the following command, please, format the Arabidopsis proteome file (Athaliana_447_Araport11.protein.fa) with DIAMOND build to get the index (Athaliana_447_Araport11.dmnd). Then, you need to fillout the configuration file PEPE.cfg (there is a template in the scripts directory).
cd ./scripts/
./PEPE.sh PEPE.cfg
This analysis depends on PEPE results (for example, FilteredReadCountsMatrix_2_3_1.txt), the goatools [https://github.com/tanghaibao/goatools] software and the reference annotation info file from Phytozome:
mkdir ../outdir/goatools_phytozome
./mkGOATOOLS.sh phytozome ../refs/Athaliana_447_Araport11.annotation_info.txt ../outdir/FilteredReadCountsMatrix_2_3_1.txt ../outdir/goatools_phytozome
... or GO association file (item2term_1) from agriGO [http://bioinfo.cau.edu.cn/agriGO/download/item2term_1]:
mkdir ../outdir/goatools_agrigo
./mkGOATOOLS.sh agrigo ./item2term_1 ../outdir/FilteredReadCountsMatrix_2_3_1.txt ../outdir/goatools_agrigo