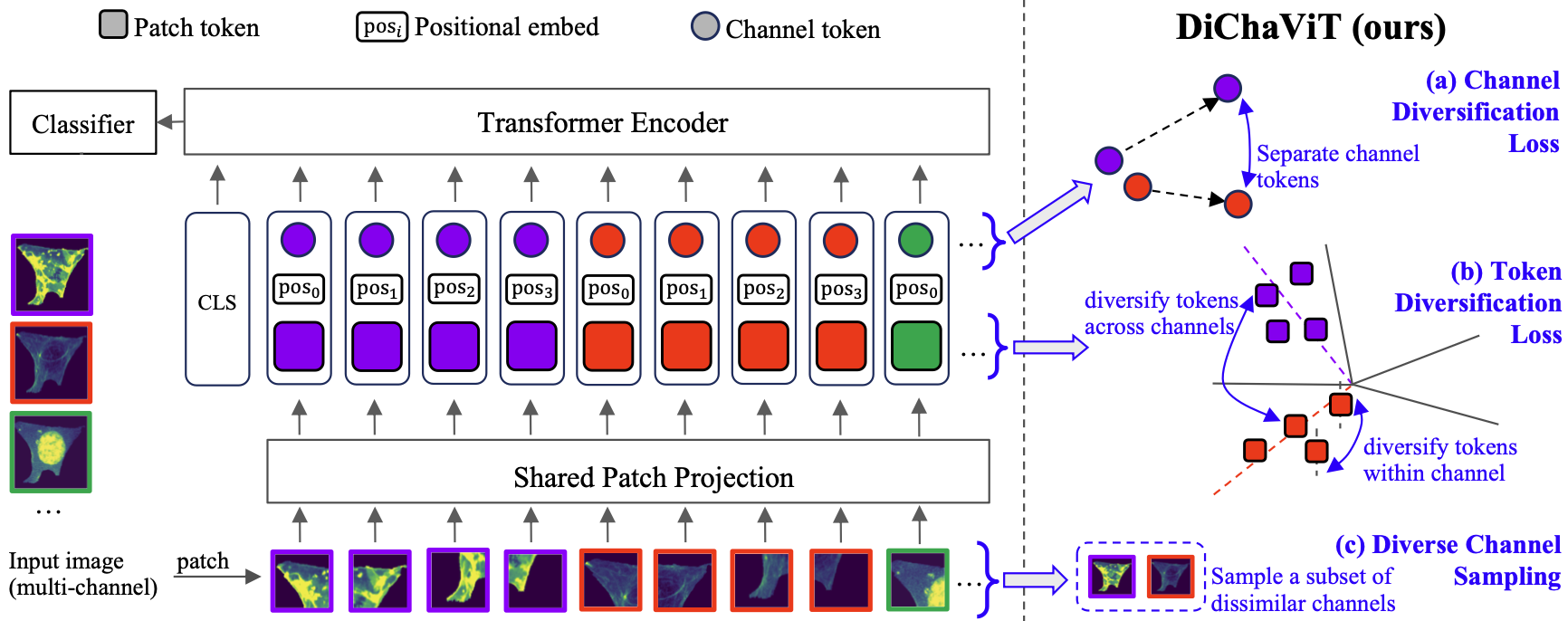
A Pytorch implementation for Diverse Channel ViT (DiChaViT) in our paper. This code was tested using Pytorch 2.4.1+cu121 and Python 3.10.
If you find our work useful, please consider citing:
@InProceedings{phamDiChaVit2024,
author = {Chau Pham and Bryan A. Plummer},
title = {Enhancing Feature Diversity Boosts Channel-Adaptive Vision Transformers},
booktitle = {Advances in Neural Information Processing Systems (NeurIPS)},
year = {2024}}
Install required packages:
conda create -n dichavit python=3.10 -y
conda activate dichavit
pip install -r requirements.txt
After downloading the following datasets, you need to update the paths in the config files configs/dataset/.
The dataset can be downloaded from https://doi.org/10.5281/zenodo.7988357
To run evaluation, we need to install the evaluation package: https://github.com/broadinstitute/MorphEm
More detail about the dataset can be found here.
You can refer to insitro's dataset repo for further details. Here's a quick overview to help you get started.
The processed data is stored in an S3 bucket as follows:
s3://insitro-research-2023-context-vit
└── jumpcp/
├── platemap_and_metadata/
├── BR00116991/
│ ├── BR00116991_A01_1_12.npy
│ ├── BR00116991_A01_1_13.npy
│ └── ...
├── BR00116993/
├── BR00117000/
├── BR00116991.pq
├── BR00116993.pq
└── BR00117000.pq
We conduct experiments on the BR00116991 dataset, which requires downloading platemap_and_metadata/, BR00116991/ folders, and BR00116991.pq.
First, you need to install AWS CLI, then run these commands in the Terminal:
aws s3 cp s3://insitro-research-2023-context-vit/jumpcp/platemap_and_metadata jumpcp/platemap_and_metadata --recursive --no-sign-request
aws s3 cp s3://insitro-research-2023-context-vit/jumpcp/BR00116991 jumpcp/BR00116991 --recursive --no-sign-request
aws s3 cp s3://insitro-research-2023-context-vit/jumpcp/BR00116991.pq jumpcp/BR00116991.pq --no-sign-request
We use the city split (version 1) of the So2Sat dataset. The dataset can be downloaded by running
wget --no-check-certificate https://dataserv.ub.tum.de/s/m1454690/download?path=%2F&files=validation.h5&downloadStartSecret=p5bjok57fil
For more detail, you can refer to So2Sat-LCZ42 repo.
In this project, we use Hydra to manage configurations. To submit a job using Hydra, you need to specify the config file. Here are some key parameters:
-m: multi-run mode (submit multiple runs with 1 job)
-cp: config folder, all config files are in `configs/`
-cn: config file name (without .yaml extension)
Parameters in the command lines will override the ones in the config file. For example, to train a DiChaViT on CHAMMI dataset:
python main.py -m -cn chammi_cfg model=dichavit ++model.enable_sample=True ++model.pretrained_model_name=small tag=test_demo dataset=morphem70k_v2_12channels ++optimizer.params.lr=0.00004 ++model.temperature=0.07 ++train.num_epochs=10 ++train.batch_size=64 ++model.new_channel_inits=[zero] ++logging.wandb.use_wandb=False ++eval.skip_eval_first_epoch=True
To reproduce the results, please refer to train_scripts.sh.
Add Wandb key: If you would like to use Wandb to keep track of experiments, add your Wandb key to .env file:
echo WANDB_API_KEY=your_wandb_key >> .env
and, change use_wandb to True in configs/logging/wandb.yaml, or set ++logging.wandb.use_wandb=True in the command line.
The model checkpoints for DiChaViT can be found here.
- ChannelViT model, and dataloaders for So2Sat and JUMP-CP are adapted from ChannelViT
- CHAMMI's baseline models, dataloader, and evaluation benchmark are from CHAMMI, MorphEm