Table of Contents
Search Stack is a web app framework enabling users to search a PostgreSQL database by a field name, with results rendered in a simple tabular view.
Included features
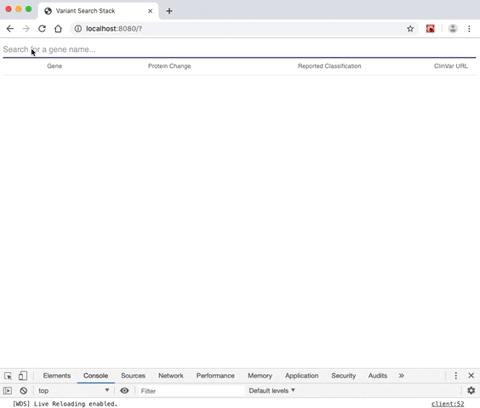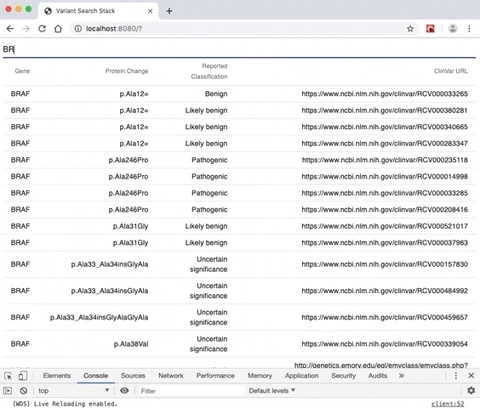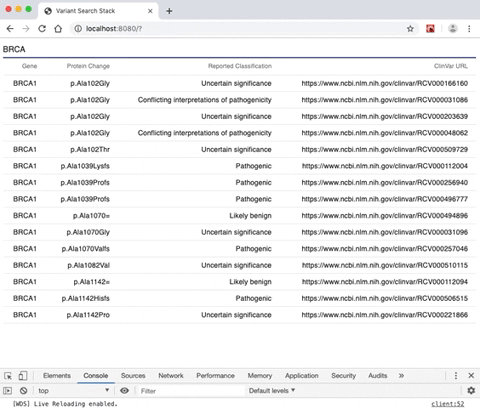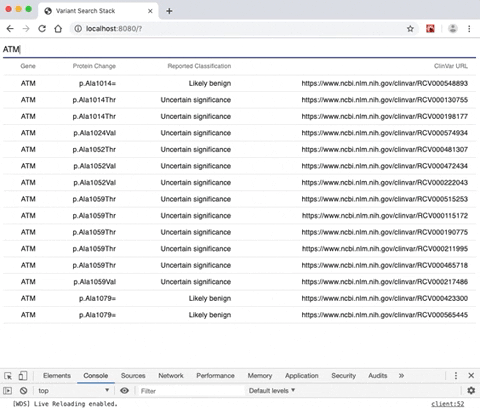
- A rendered search result from a gene selection input allowing the user to view a list of genomic variants alongside variant-specific attributes.
- Throttle-debounced XHR requesting thus allowing for fewer GETS to database server.
- Search is throttled query is fewer than 5 characters
- Search is debounced over 5 characters
- A RESTful endpoint serving gene-name query functionality.
- Input and Table components from React Material-UI provide a minimal and functional inferface.
- Create a virtual conda (Python 3) environment called
search_stack-envwith Python and pip
➜ conda create --name search_stack-env python=3.6 pip
➜ source activate search_stack-env
(search_stack-env) ➜- Clone the repository and install package requirements.
(search_stack-env) ➜ git clone https://github.com/foadgr/search_stack.git
(search_stack-env) ➜ cd search_stack
(search_stack-env) ➜ pip install -r requirements.txtA PostgreSQL database image can be pulled from Docker.
sudo docker run --name app_db -p 5433:5432 \
-e POSTGRES_PASSWORD= \
-d fgreenInstall the latest version of PostgREST, make the package executable (optional), and run the server based on the server configuration in config/setup/db.conf.
# Download from https://github.com/PostgREST/postgrest/releases/latest
(search_stack-env) ➜ tar xfJ postgrest-v6.0.2-osx.tar.xz
# As an executable
(search_stack-env) ➜ cp postgrest /usr/local/bin
(search_stack-env) ➜ postgrest config/setup/db.conf
# Or run outside of bin
(search_stack-env) ➜ ./postgrest config/setup/db.conf
# You should see the following output
>>> Listening on port 3000
>>> Attempting to connect to the database...
>>> Connection successfulInstall the React client and run the build.
npm install
npm run buildIf the build is successful, run the server.
npm startThe client can be viewed on port 8080.
Extract genomic variatns source data from web source, data cleaning, and load to app_db genomic variants data from web source.
(search_stack-env) ➜ python search_stack/insert.pyRunning insert.py also initiates the necessary API schema within the Postgres app_db database, which includes the main store table, a filtered view, alongside a table index designed with the purpose of efficient gene lookup. A regular B-Tree index only a prefix of the gene will help save space of the index size. Structuring the index with the varchar_pattern_ops operator class will help speed up the gene ~* {user_input}% query.
-- Create the B-Tree index
CREATE INDEX ON api.variants (substr(gene, 1, 4) varchar_pattern_ops);
--This index will work alongside the main REST endpoint in the search query
SELECT gene FROM api.variants WHERE substr(gene, 1, 4) ~* '{user input}%'