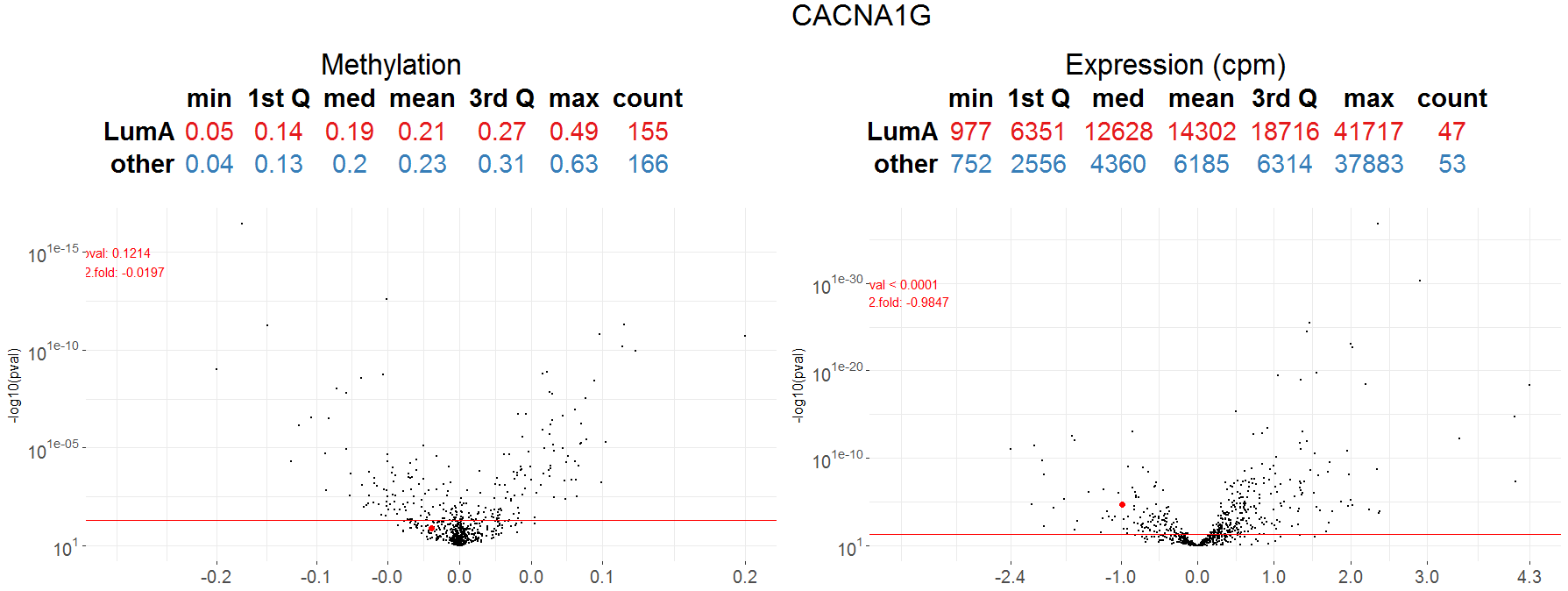
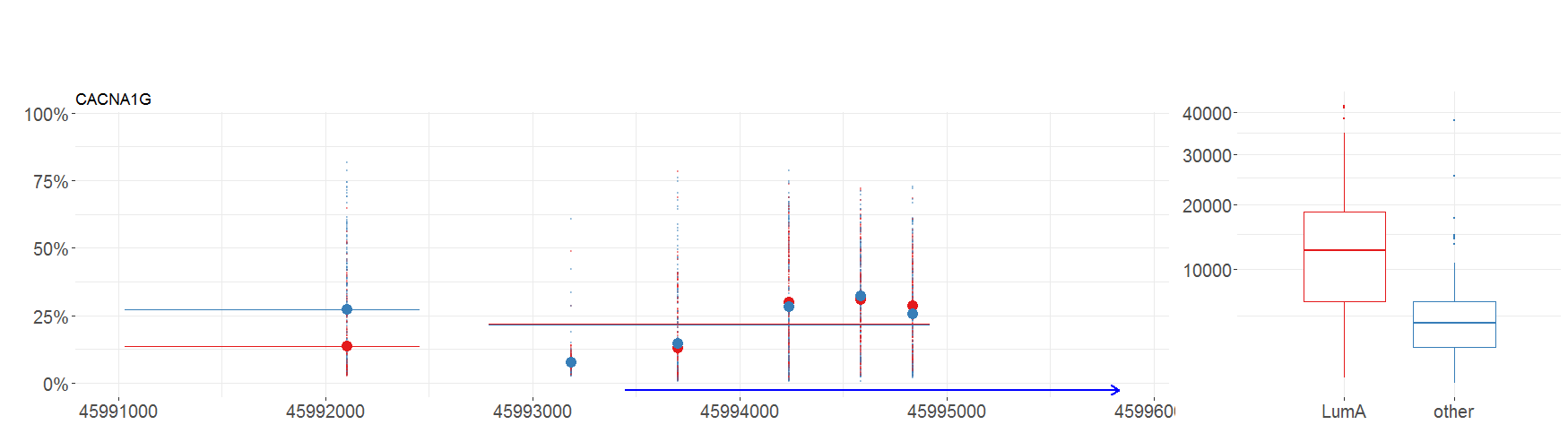
Machine Learning for Genetic Signatures
Package for analyzing genes expression and CpG probes methylation.
Our package uses a few packages from Bioconductor. To install them, start R and enter
## try http:// if https:// URLs are not supported
source("https://bioconductor.org/biocLite.R")
biocLite("DESeq")
## try http:// if https:// URLs are not supported
source("https://bioconductor.org/biocLite.R")
biocLite("DESeq2")
## try http:// if https:// URLs are not supported
source("https://bioconductor.org/biocLite.R")
biocLite("limma")
## try http:// if https:// URLs are not supported
source("https://bioconductor.org/biocLite.R")
biocLite("edgeR")
## try http:// if https:// URLs are not supported
source("https://bioconductor.org/biocLite.R")
biocLite("methyAnalysis")
## try http:// if https:// URLs are not supported
source("https://bioconductor.org/biocLite.R")
biocLite("TxDb.Hsapiens.UCSC.hg18.knownGene")
## try http:// if https:// URLs are not supported
source("https://bioconductor.org/biocLite.R")
biocLite("org.Hs.eg.db")
To install this package, start R and enter:
devtools::install_github("geneticsMiNIng/MLGenSig/MLExpResso")
In order to run examples you shall install MLExpRessoData.
devtools::install_github("geneticsMiNIng/MLGenSigdata/MLExpRessodata")
- Full BRCA mRNAseq data
- Full BRCA mRNAseq data with subtypes
- BRCA clinical data with parameters
- BRCA clinical data w/o parameters
Work on this package was financially supported by the 'NCN Opus grant 2016/21/B/ST6/02176'.