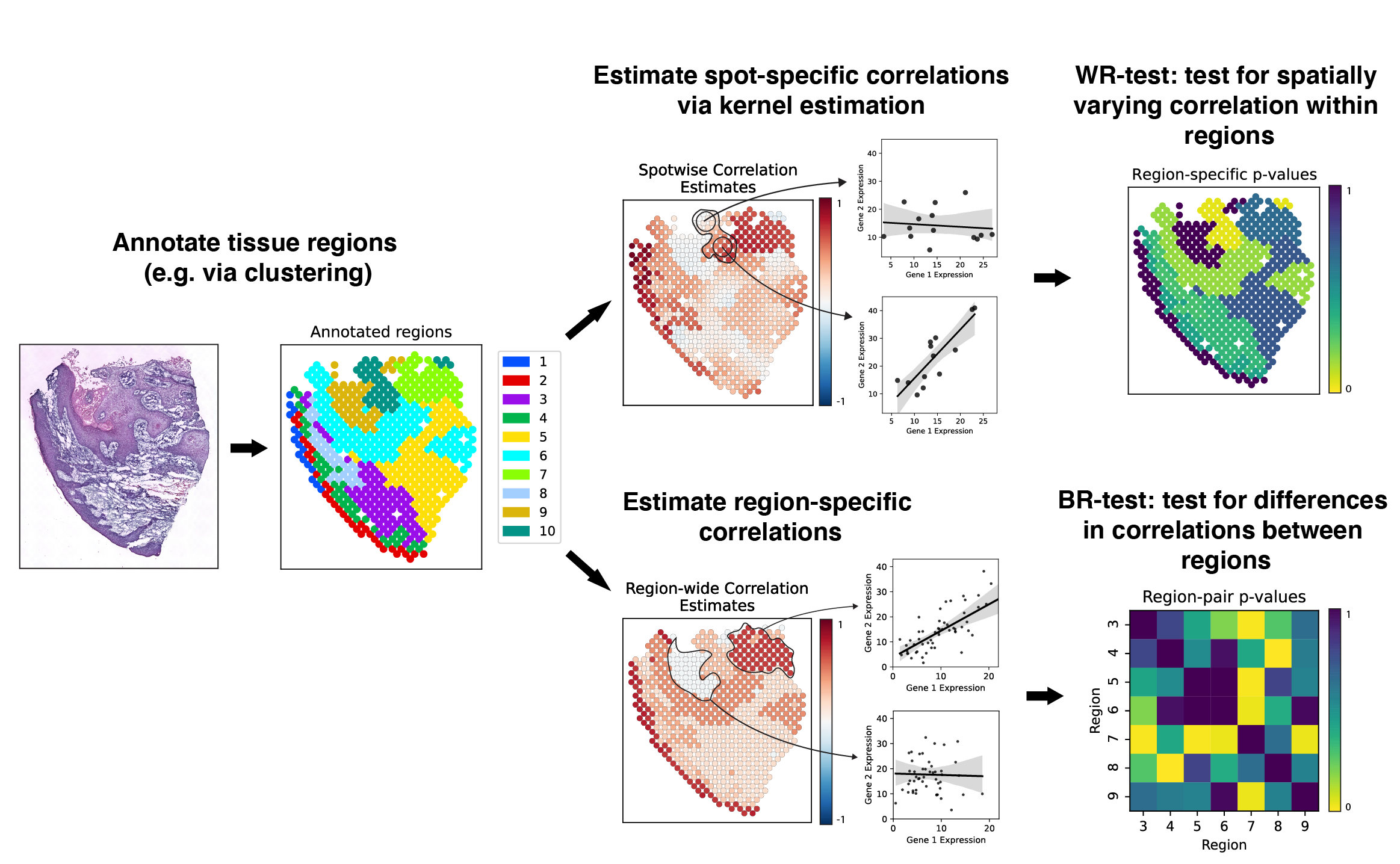
SpatialCorr is a set of statistical methods for identifying genes whose correlation structure changes across a spatial transcriptomics sample. Along with a set of statistical tests, SpatialCorr also offers a number of methods for visualizing spatially varying correlation.
Here is a schematic overview of the analyses performed by SpatialCorr:
For more details regarding the underlying method, see the paper:
Bernstein, M.N., Ni, Z., Prasad, A., Brown, J., Mohanty, C., Stewart, R., Newton, M.A., Kendziorski, C. (2022). SpatialCorr: Identifying gene sets with spatially varying correlation structure. Cell Reports Methods.
To install SpatialCorr using Pip, run the following command:
pip install spatialcorr
For SpatialCorr's API manual, please visit the documentation.
For a tutorial on running SpatialCorr, please see the tutorial. This tutorial can also be run via Google Colab.