-
Notifications
You must be signed in to change notification settings - Fork 9
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Merge pull request #67 from usegalaxy-eu/pathogfair
Add thread for PathoGFAIR preprint
- Loading branch information
Showing
2 changed files
with
59 additions
and
0 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,59 @@ | ||
| --- | ||
| media: | ||
| - mastodon-eu-freiburg | ||
| - matrix-eu-announce | ||
| - linkedin-galaxyproject | ||
|
|
||
| mentions: | ||
| mastodon-eu-freiburg: | ||
| - [email protected] | ||
|
|
||
| hashtags: | ||
| mastodon-eu-freiburg: | ||
| - metagenomics | ||
| - pathogen | ||
| - UseGalaxy | ||
|
|
||
| --- | ||
|
|
||
| **🚀 Exciting news in pathogen detection!** | ||
| Our new PathoGFAIR preprint introduces FAIR and adaptable (meta)genomics workflows for detecting and tracking foodborne pathogens. This project started nearly three years ago as a collaboration with Biolytix, a swiss SME. 🌟 | ||
|
|
||
| 📝 [Preprint](https://www.biorxiv.org/content/10.1101/2024.06.26.600753v1) | ||
| 🔗 [Blogpost](https://galaxyproject.org/news/2024-07-08-pathogfair-preprint/) | ||
|
|
||
|
|
||
| **🏁 Project Origins** | ||
| The journey began with funding from EOSC-Life for Industry Collaboration to modernize and validate Biolytix's bioinformatic pipelines. The goal? To make pathogen detection workflows accessible and scalable via the Galaxy platform. 🌐 | ||
|
|
||
| 🔗 [Project Background](https://galaxyproject.org/news/2021-12-08-pathogen-detection-eosc-life-grant/) | ||
|
|
||
|
|
||
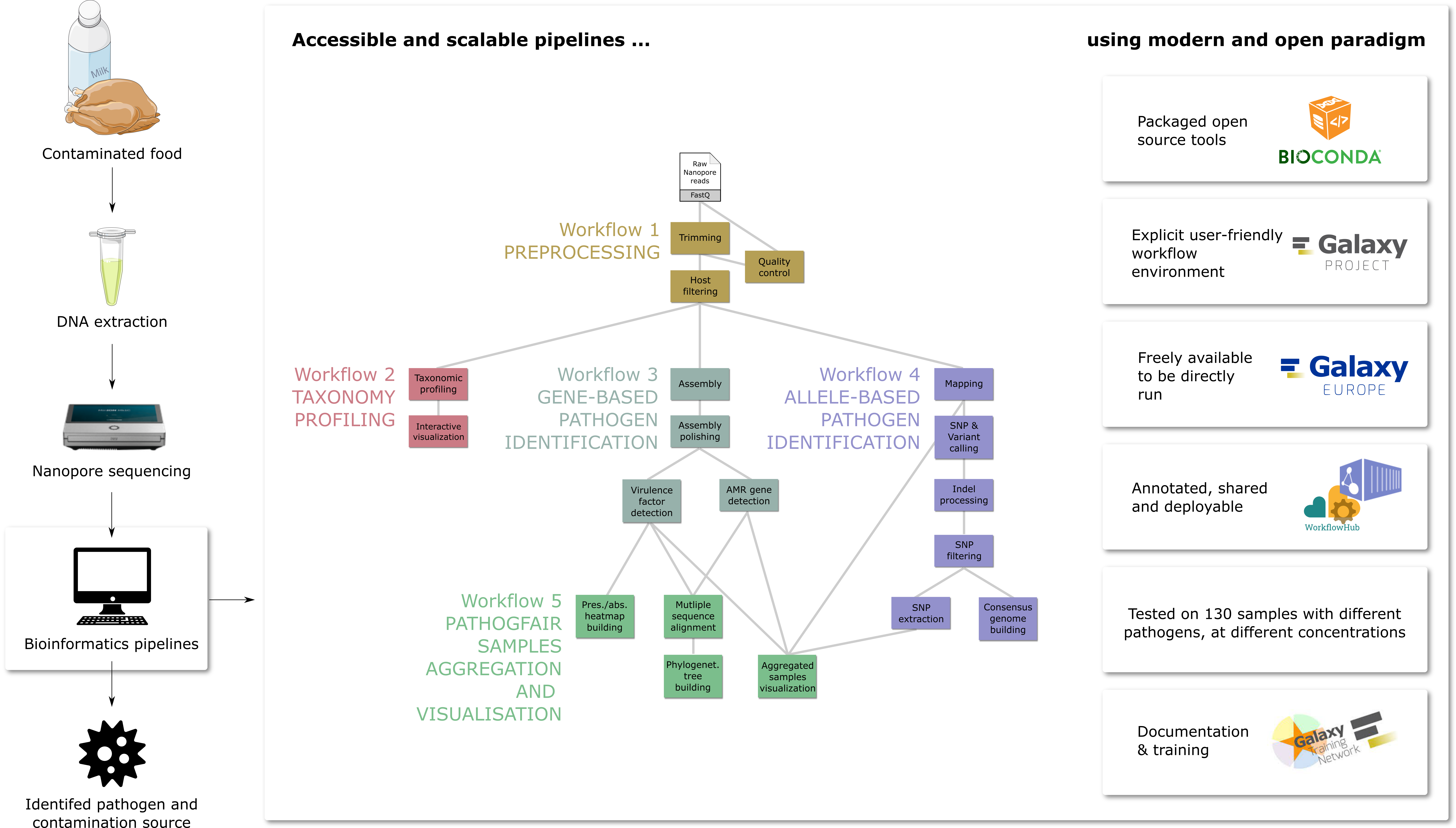
| **🔍 What's in PathoGFAIR?** | ||
| PathoGFAIR consists of 5 workflows: | ||
| - **Preprocessing** | ||
| - **Taxonomy Profiling and Visualization** | ||
| - **Gene-based Pathogen Identification** with assembly, AMR and VF gene detection | ||
| - **Allele-based Pathogen Identification** with variant calling | ||
| - **PathoGFAIR Samples Aggregation and Visualisation** including phylogenetic analysis | ||
|
|
||
| 🔗 [Workflows](https://usegalaxy-eu.github.io/PathoGFAIR/#how-to-find-pathogfair-workflows) | ||
|
|
||
|
|
||
| **🌟 Embracing FAIR Principles** | ||
| These workflows adhere to FAIR (Findable, Accessible, Interoperable, Reusable) principles, ensuring they are easy to find (via WorkflowHub and Dockstore), share, adapt, and integrate with other data and tools. This enhances collaboration and transparency in pathogen research. | ||
|
|
||
|  | ||
|
|
||
|
|
||
| **📚 Training Material Available** | ||
| To support users, a comprehensive tutorial has been developed. It guides users through the workflows, ensuring they can effectively utilize the tools for their research and public health initiatives. | ||
|
|
||
| 🔗 [Tutorial](https://training.galaxyproject.org/training-material/topics/microbiome/tutorials/pathogen-detection-from-nanopore-foodborne-data/tutorial.html) | ||
|
|
||
|
|
||
| **🌍 Impact** | ||
| PathoGFAIR is set to improve pathogen tracking, providing robust tools for researchers and public health officials worldwide. By improving detection capabilities, we can better protect public health. 🌎🔬 | ||
|
|
||
| 📝 [Preprint](https://www.biorxiv.org/content/10.1101/2024.06.26.600753v1) | ||
| 🔗 [Blogpost](https://galaxyproject.org/news/2024-07-08-pathogfair-preprint/) |
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.