-
Notifications
You must be signed in to change notification settings - Fork 8
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Posting PathoGFAIR preprint to bluesky #72
Conversation
This comment was marked as outdated.
This comment was marked as outdated.
|
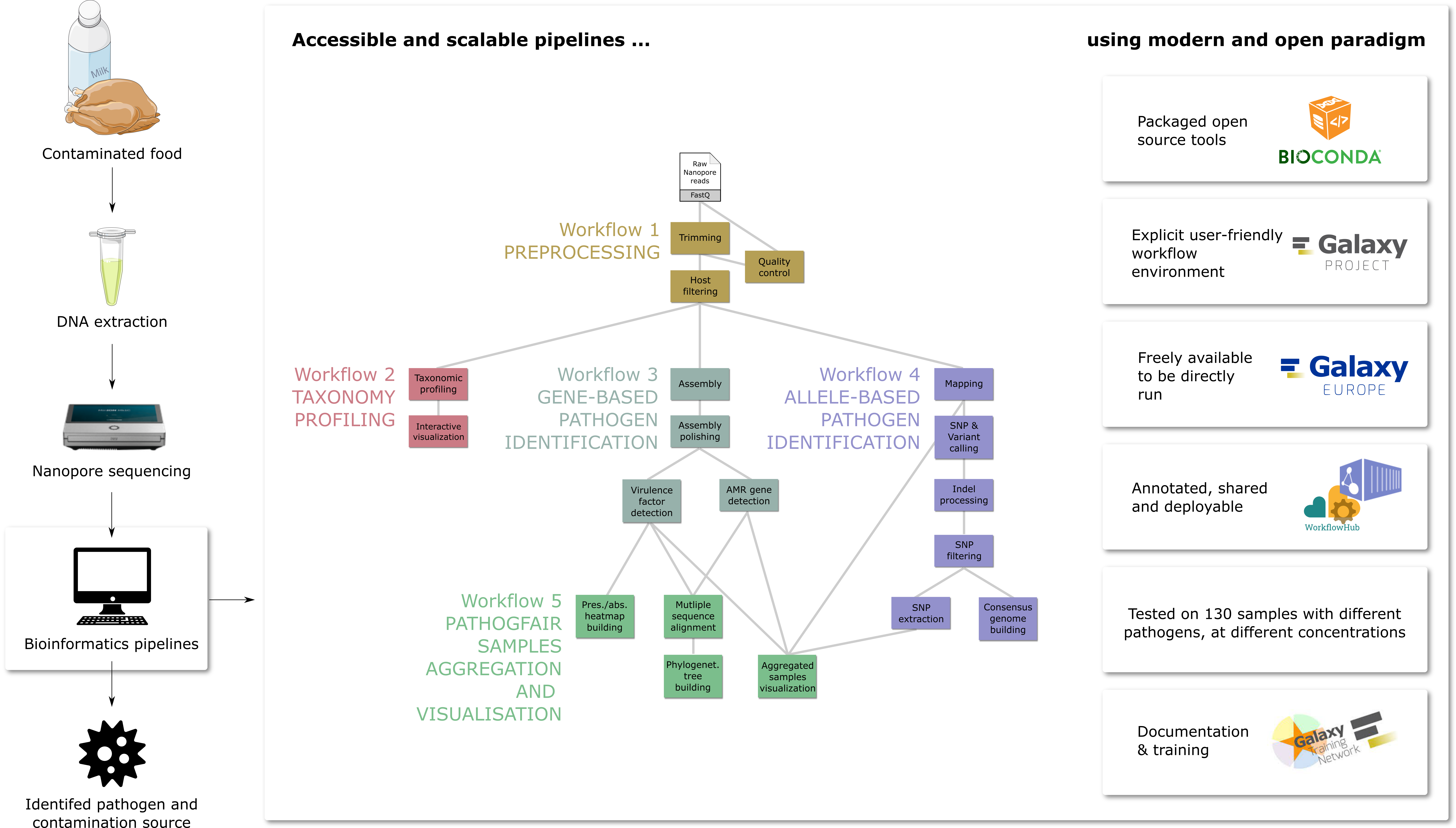
👋 Hello! I'm your friendly social media assistant. Below are the previews of this post: Skipping post to mastodon-eu-freiburg, matrix-eu-announce, linkedin-galaxyproject. because it was already posted. bluesky-galaxyproject🚀 Exciting news in pathogen detection! 📝 Preprint: (1/12) https://www.biorxiv.org/content/10.1101/2024.06.26.600753v1 🏁 Project Origins 🔗 Project Background: (3/12) https://galaxyproject.org/news/2021-12-08-pathogen-detection-eosc-life-grant/ (4/12) 🔍 What's in PathoGFAIR?
Visualisation including phylogenetic analysis 🔗 Workflows: https://usegalaxy-eu.github.io/PathoGFAIR/#how-to-find-pathogfair-workflows (6/12) 🌟 Embracing FAIR Principles pathogen research. (8/12) 📚 Training Material Available 🔗 Tutorial: (9/12) 🌍 Impact 📝 Preprint: https://www.biorxiv.org/content/10.1101/2024.06.26.600753v1 Blogpost: https://galaxyproject.org/news/2024-07-08-pathogfair-preprint/
|
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Hmm, why is this getting split in such a weird way?
|
The maximum Bluesky content for each post is 300 characters. It also applied the manual separation, making it look like this. |
|
Ah, I see. |
|
Should we create another file for posting to Bluesky? #76 |
No description provided.