-
Notifications
You must be signed in to change notification settings - Fork 1
DTX1
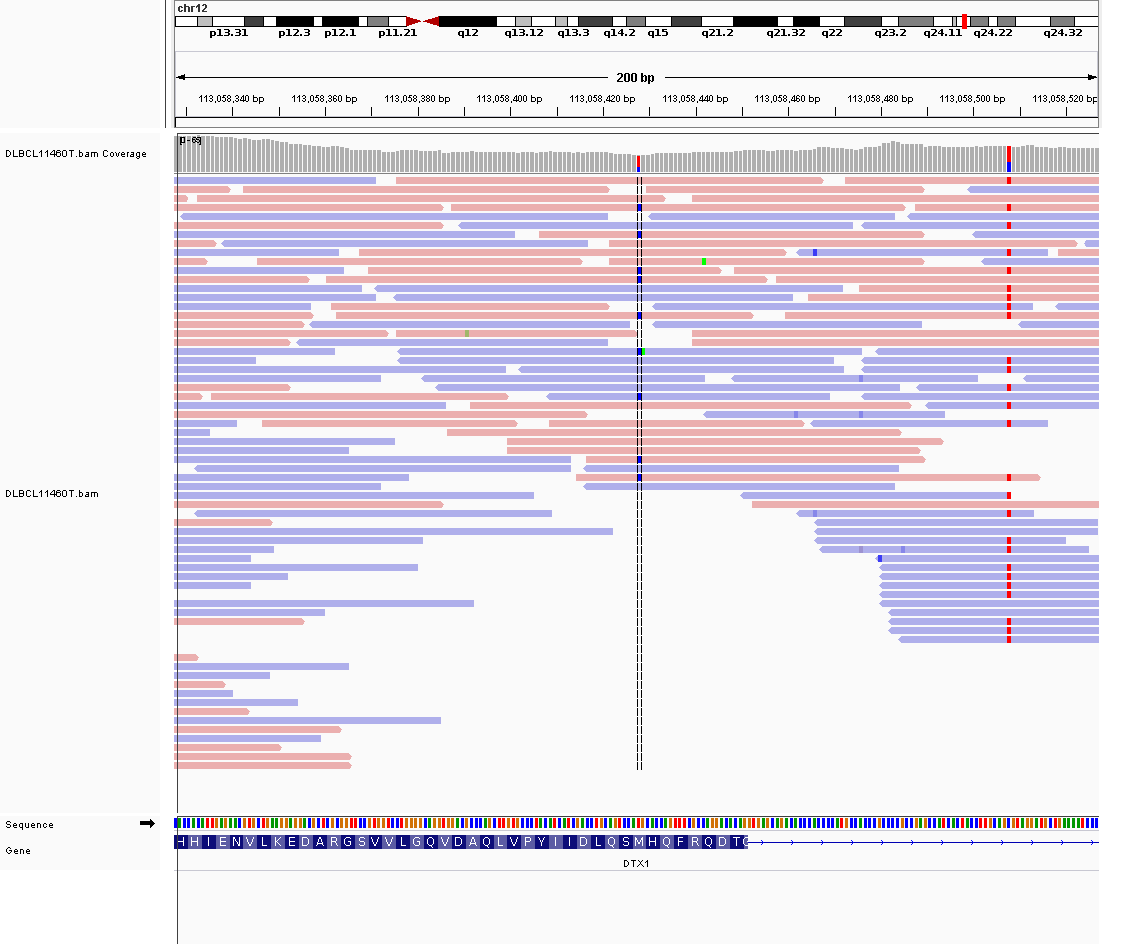
Mutations in the DTX1 gene, which encodes the E3 ubiquitin ligase Deltex 1, have been identified in various B-cell lymphomas, including diffuse large B-cell lymphoma (DLBCL). DTX1 is one of a number of genes affected by aberrant somatic hypermutation in B-cell lymphomas, which complicates the interpretation of mutations at this locus. These mutations are associated with the BN2 genetic subgroup of DLBCL. There are numerous mutation hotspots in this gene with some leading to a truncated protein. DTX1 functions as a negative regulator of the Notch signaling pathway. Some DTX1 mutations impair its function, thereby dysregulating Notch signaling, which is crucial for normal B-cell development and function. Due to minimal support in the original primary data and very few mutations reported in subsequent BL studies, this gene is very unlikely to be relevant in BL.
%%{init: { 'logLevel': 'debug', 'theme': 'dark' } }%%
timeline
title Publication timing
2012-08-27 : Rossi : MZL
2012-12-01 : Love : BL
2013-01-01 : Zhang : DLBCL
2018-04-12 : Schmitz : DLBCL
2018-05-01 : Chapuy : DLBCL
2023-11-15 : Gomez : PMBL
| Entity | Tier | Description |
|---|---|---|
| 1 | high-confidence PMBL/cHL/GZL gene[@gomezUltraDeepSequencingReveals2023] | |
| 1 | high-confidence MZL gene[@rossiCodingGenomeSplenic2012c] | |
| 3 | Retired, Failed QC[@loveGeneticLandscapeMutations2012] | |
| 1 | aSHM target and high-confidence DLBCL gene [@zhangGeneticHeterogeneityDiffuse2013; @schmitzGeneticsPathogenesisDiffuse2018a; @chapuyMolecularSubtypesDiffuse2018b] | |
| 1 | aSHM target and high-confidence FL gene |
| Entity | source | frequency (%) |
|---|---|---|
| DLBCL | GAMBL genomes | 10.71 |
| DLBCL | Schmitz cohort | 13.62 |
| DLBCL | Reddy cohort | 5.91 |
| DLBCL | Chapuy cohort | 11.54 |
| FL | GAMBL genomes | 5.54 |
| BL | GAMBL genomes+capture | 5.54 |
| BL | Thomas cohort | 5.90 |
| BL | Panea cohort | 8.90 |
| Entity | aSHM | Significant selection | dN/dS (missense) | dN/dS (nonsense) |
|---|---|---|---|---|
| DLBCL | Yes | Yes | 2.228 | 4.244 |
| FL | Yes | No | 6.305 | 0.000 |
| BL | Yes | No | 5.556 | 14.253 |
| chr_name | hg19_start | hg19_end | region | regulatory_comment |
|---|---|---|---|---|
| chr12 | 113492311 | 113497546 | TSS | NA |
| Chromosome | Coordinate (hg19) | ref>alt | HGVSp |
|---|---|---|---|
| chr12 | 113496076 | G>A | V27M |
| chr12 | 113496081 | G>A | W28* |
| chr12 | 113496082 | G>A | E29K |
| chr12 | 113496083 | A>G | E29G |
| chr12 | 113496085 | T>G | W30G |
| chr12 | 113496087 | G>A | W30* |
| chr12 | 113496089 | T>A | L31Q |
| chr12 | 113496089 | T>C | L31P |
| chr12 | 113496096 | G>C | E33D |
| chr12 | 113496097 | C>T | H34Y |
| chr12 | 113496112 | C>T | P39S |
| chr12 | 113496115 | T>C | Y40H |
| chr12 | 113496116 | A>C | Y40S |
| chr12 | 113496116 | A>G | Y40C |
| chr12 | 113496117 | C>G | Y40* |
| chr12 | 113496121 | G>C | A42P |
| chr12 | 113496130 | T>A | C45S |
| chr12 | 113496132 | C>A | C45* |
| chr12 | 113496133 | C>T | H46Y |
| chr12 | 113496135 | C>A | H46Q |
| chr12 | 113496139 | A>T | I48F |
| chr12 | 113496140 | T>A | I48N |
| chr12 | 113496141 | T>G | I48M |
| chr12 | 113496148 | G>A | V51M |
| chr12 | 113496155 | A>T | K53M |
| chr12 | 113496156 | G>C | K53N |
| chr12 | 113496159 | G>C | E54D |
| chr12 | 113496162 | CG>AA | DA55ET |
| chr12 | 113496170 | G>A | G58D |
| chr12 | 113496173 | C>A | S59Y |
| chr12 | 113496173 | C>T | S59F |
| chr12 | 113496175 | G>A | V60M |
| chr12 | 113496191 | T>C | V65A |
| chr12 | 113496191 | T>G | V65G |
| chr12 | 113496196 | G>A | A67T |
| chr12 | 113496197 | C>T | A67V |
| chr12 | 113496201 | G>C | Q68H |
| chr12 | 113496202 | C>T | L69F |
| chr12 | 113496205 | G>C | V70L |
| chr12 | 113496208 | C>G | P71A |
| chr12 | 113496208 | C>T | P71S |
| chr12 | 113496210 | CT>TC | Y72H |
| chr12 | 113496212 | A>G | Y72C |
| chr12 | 113496213 | C>A | Y72* |
View coding variants in ProteinPaint hg19 or hg38
View all variants in GenomePaint hg19 or hg38
 Rating
★ ☆ ☆ ☆ ☆
Rating
★ ☆ ☆ ☆ ☆
 Rating
★ ★ ★ ★ ☆
Rating
★ ★ ★ ★ ☆


Disclaimer
The content in these pages has been populated, in part, by an automated process. Although we have scrutinized every page to ensure accuracy, errors will inevitably exist. If you find an error please report it as an issue and we will address it.
In particular, let us know if you feel that an important citation is missing or if a paper has been cited incorrectly.
License
This work is licensed under a Creative Commons Attribution 4.0 International License.

You are free to:
Share — copy and redistribute the material in any medium or format for any purpose, even commercially.
Adapt — remix, transform, and build upon the material for any purpose, even commercially. The licensor cannot revoke these freedoms as long as you follow the license terms.